filmov
tv
Eukaryotic mRNA Processing; Spliceosome

Показать описание
Need help preparing for the Biology section of the MCAT? MedSchoolCoach expert, Ken Tao, will teach everything you need to know about Eukaryotic mRNA Processing and Spliceosome of Transcription. Watch this video to get all the MCAT study tips you need to do well on this section of the exam!
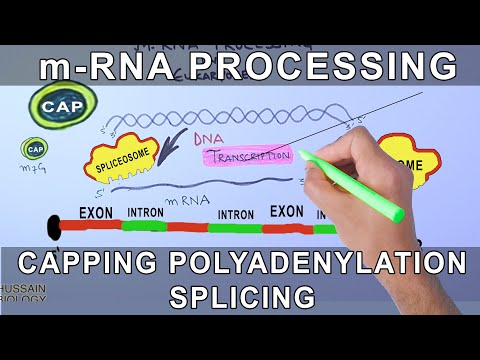
The original RNA molecule, synthesized by RNA polymerase, will undergo several modifications before forming mRNA. These processes are referred to as eukaryotic mRNA processing. The original mRNA molecule that is produced is called pre-mRNA. Only until the pre-mRNA undergoes mRNA processing, will it become mRNA. The location of eukaryotic mRNA processing is in the nucleus of the cell. In this way, any mRNA that has been released from the nucleus into the cytoplasm has undergone these modifications.
Eukaryotic mRNA Modifications
Eukaryotic mRNA processing consists of three modifications to the RNA molecule: The addition of a 5’ cap, a 3’ poly-A tail, and RNA splicing. The 5’ cap refers to the addition of a methyl-guanine at the 5’ end of the pre-mRNA. The poly-A tail refers to polyadenylation or the addition of a chain of adenosine and RNA molecules at the 3’ end of the pre-mRNA.
The purpose of both the 5’ cap and the 3’ poly-A tail is to protect the RNA from degradation and to promote translation. RNA is not a very stable molecule, so without these particular modifications, it would degrade easily and quickly. Since mRNA is to be translated into proteins, it is vital to ensure its viability. The last modification of eukaryotic mRNA processing is RNA splicing. The pre-mRNA molecule consists of both nucleotides that will code for the protein of interest and nucleotides that will not. Splicing is the process by which the noncoding regions, called introns, are spliced out or removed. When the introns are removed, what is left are the coding regions of RNA, known as exons.
Alternative Splicing
RNA splicing does not occur in the same fashion every time, as the pre-mRNA molecule can be spliced in multiple ways. This variety in splicing is referred to as alternative splicing. Alternative splicing is essential because it creates a more efficient system of protein production. It is a process that allows for the formation of different versions of mature mRNA, and therefore different protein products, from only a single gene. Instead of needing new gene sequences to create different protein products, a single gene sequence is sufficient enough to create multiple protein products.
What is interesting to note is that alternative mRNA splicing has been shown to alter the diversity of certain diseases, such as cancer. While at first glance, this may seem problematic, researchers believe that the broader and more varied the cancer, the more targets for cancer immunotherapy. To date, the vast array of alternative splicing-derived cancer targets has been mainly unexplored. Perhaps they can offer an effective way to treat previously untreatable cancers.
Spliceosomes
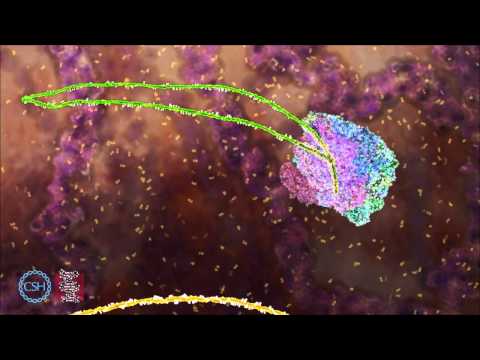
The spliceosome is a very large RNA and protein complex located in the nucleus of the cell, which acts as the machinery used for RNA splicing. Each spliceosome complex is made up of several types of small nuclear RNA, or snRNA, and protein factors. The combination of the snRNA and protein factors create what are known as small nuclear ribonucleoproteins, or snRNPs. The snRNPs then bind to pre-mRNA molecules creating a new spliceosome complex.
MEDSCHOOLCOACH
To watch more MCAT video tutorials like this and have access to study scheduling, progress tracking, flashcard and question bank, download MCAT Prep by MedSchoolCoach
#medschoolcoach #MCATprep #MCATstudytools
The original RNA molecule, synthesized by RNA polymerase, will undergo several modifications before forming mRNA. These processes are referred to as eukaryotic mRNA processing. The original mRNA molecule that is produced is called pre-mRNA. Only until the pre-mRNA undergoes mRNA processing, will it become mRNA. The location of eukaryotic mRNA processing is in the nucleus of the cell. In this way, any mRNA that has been released from the nucleus into the cytoplasm has undergone these modifications.
Eukaryotic mRNA Modifications
Eukaryotic mRNA processing consists of three modifications to the RNA molecule: The addition of a 5’ cap, a 3’ poly-A tail, and RNA splicing. The 5’ cap refers to the addition of a methyl-guanine at the 5’ end of the pre-mRNA. The poly-A tail refers to polyadenylation or the addition of a chain of adenosine and RNA molecules at the 3’ end of the pre-mRNA.
The purpose of both the 5’ cap and the 3’ poly-A tail is to protect the RNA from degradation and to promote translation. RNA is not a very stable molecule, so without these particular modifications, it would degrade easily and quickly. Since mRNA is to be translated into proteins, it is vital to ensure its viability. The last modification of eukaryotic mRNA processing is RNA splicing. The pre-mRNA molecule consists of both nucleotides that will code for the protein of interest and nucleotides that will not. Splicing is the process by which the noncoding regions, called introns, are spliced out or removed. When the introns are removed, what is left are the coding regions of RNA, known as exons.
Alternative Splicing
RNA splicing does not occur in the same fashion every time, as the pre-mRNA molecule can be spliced in multiple ways. This variety in splicing is referred to as alternative splicing. Alternative splicing is essential because it creates a more efficient system of protein production. It is a process that allows for the formation of different versions of mature mRNA, and therefore different protein products, from only a single gene. Instead of needing new gene sequences to create different protein products, a single gene sequence is sufficient enough to create multiple protein products.
What is interesting to note is that alternative mRNA splicing has been shown to alter the diversity of certain diseases, such as cancer. While at first glance, this may seem problematic, researchers believe that the broader and more varied the cancer, the more targets for cancer immunotherapy. To date, the vast array of alternative splicing-derived cancer targets has been mainly unexplored. Perhaps they can offer an effective way to treat previously untreatable cancers.
Spliceosomes
The spliceosome is a very large RNA and protein complex located in the nucleus of the cell, which acts as the machinery used for RNA splicing. Each spliceosome complex is made up of several types of small nuclear RNA, or snRNA, and protein factors. The combination of the snRNA and protein factors create what are known as small nuclear ribonucleoproteins, or snRNPs. The snRNPs then bind to pre-mRNA molecules creating a new spliceosome complex.
MEDSCHOOLCOACH
To watch more MCAT video tutorials like this and have access to study scheduling, progress tracking, flashcard and question bank, download MCAT Prep by MedSchoolCoach
#medschoolcoach #MCATprep #MCATstudytools
 0:04:45
0:04:45
 0:03:17
0:03:17
 0:06:53
0:06:53
 0:01:38
0:01:38
 0:08:05
0:08:05
 0:00:54
0:00:54
 0:04:48
0:04:48
 0:04:55
0:04:55
 0:11:27
0:11:27
 0:03:24
0:03:24
 0:23:32
0:23:32
 0:09:33
0:09:33
 0:02:51
0:02:51
 0:08:02
0:08:02
 0:07:04
0:07:04
 0:19:03
0:19:03
 0:08:06
0:08:06
 0:19:06
0:19:06
 0:11:22
0:11:22
 0:20:58
0:20:58
 0:15:19
0:15:19
 0:05:30
0:05:30
 0:21:26
0:21:26
 0:22:13
0:22:13