filmov
tv
How I analyze RNA Seq Gene Expression data using DESeq2

Показать описание
Book a Session (One on One)
#ATTENTION!!!!!!!!!!!!!!!!!!
Support my work
Get more bioinformatics tutorials on Patreon
Subscribe to my channels
Reach out
ATTENTION!!!!!!!!!!!!!!!!!!
Chapters
00:00 Intro
02:43 Load R libraries
03:38 Load the dataset
07:11 Set the factors
08:19 Create deseq DESeqDataSetFromMatrix() dds object and import coutn data and sample information
11:37 Filter genes
14:08 Perform statistical tests to identify differentially expressed genes (result dds)
18:34 Order the deseq_result
20:48 Make queries
24:20 Filter differentially expressed genes.
28:45 Save deseq result
31:28 Examine the output files
33:06 Visualize deseq result
33:24 Plot dispersions
35:30 PCA Plot
43:08 Heatmaps
44:11 Heatmap of sample-sample distance matrix using pheatmap
52:37 Heatmap of log transformed normalized counts using pheatmap
1:02:18 Heatmap Z-scores
1:06:51 MA Plot
01:11:37 Volcano plot using ggplot
01:16:31 Next Tutorial
DESeq2 R Tutorial
Transcriptomics
RNA-Seq Analysis
RNASeq
RNA Seq
Gene expression
RNA Seq Analysis - Bioinformatics Project
DESeq2 tutorial for gene expression analysis
bioinformatics for beginners
bioinformatics tutorial
bioinformatics course
Bioinformatics Project
#bioinformaticsforbeginners #bioinformatics #bioinformática #linuxforbeginners #linuxinformaticscoach
#bioinformaticsforbeginners #bioinformatics #bioinformática #linuxforbeginners #linux #rna #rnaseq
Комментарии
 0:36:09
0:36:09
 0:57:35
0:57:35
 0:18:26
0:18:26
 1:20:28
1:20:28
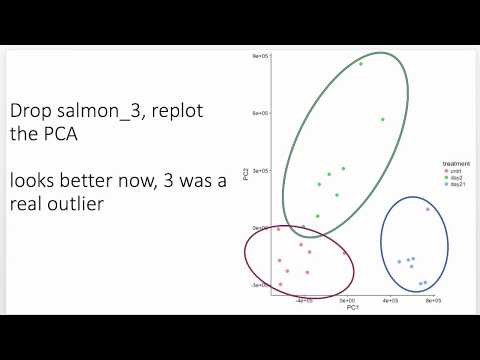 0:11:32
0:11:32
 0:14:22
0:14:22
 0:31:11
0:31:11
 0:05:50
0:05:50
 0:39:53
0:39:53
 0:04:17
0:04:17
 0:04:25
0:04:25
 0:58:58
0:58:58
 0:05:15
0:05:15
 0:43:32
0:43:32
 0:33:29
0:33:29
 0:02:35
0:02:35
 1:18:01
1:18:01
 0:15:07
0:15:07
 0:12:53
0:12:53
 0:24:37
0:24:37
 0:05:14
0:05:14
 0:01:29
0:01:29
 1:17:39
1:17:39
 0:12:11
0:12:11