filmov
tv
RNA Splicing by the Spliceosome: Supplemental Video 5

Показать описание
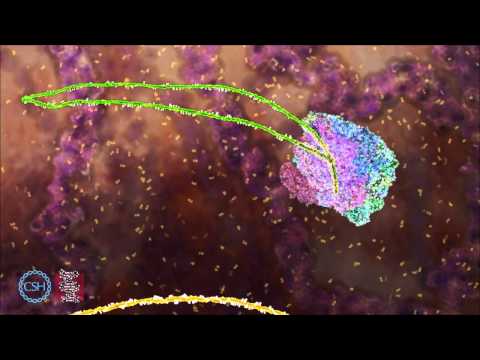
Shown: The branching reaction, showing the transition from Bact complex to C complex via B* complex. Structures derive from PDB codes 5GM6 for yeast Bact complex (82), 6J6N and 6J6Q for yeast B* complex (91), and 5LJ5 for yeast C complex (29). B* and C complex structures were modified according to a 2.8 Å resolution cryo-EM map of yeast C complex (M.E. Wilkinson, unpublished). Data not shown from other important structures (30, 81). The B* structures were used to animate docking of the branch helix and the effects of Cwc25. Transcription of narration: “In Bact complex, the branching reaction is inhibited because the 5′SS is protected by Cwc24, and the branch point is sequestered inside SF3b. To relieve this inhibition, the helicase Prp2 pulls the intron near the 3′SS. By an unknown mechanism, this triggers loss of the RES complex, SF3b, and Cwc24. The exposed branch helix can now start to dock into the active site. Docking is accompanied by a large conformational change to B* complex. For branching chemistry to occur, the step I factors Yju2 and Isy1 further dock the branch helix into the active site. Cwc25 then penetrates the branch helix to ultimately promote chemistry. The product of branching is cleavage at the 5′SS and formation of a 2′–5′ link between the 5′SS and branch point. The resultant spliceosome after branching is called C complex.” Video used with permission from the MRC Laboratory of Molecular Biology, Cambridge, UK.
 0:01:38
0:01:38
 0:06:53
0:06:53
 0:09:33
0:09:33
 0:01:03
0:01:03
 0:08:05
0:08:05
 0:10:28
0:10:28
 0:02:01
0:02:01
 0:04:55
0:04:55
 0:01:05
0:01:05
 0:04:16
0:04:16
 0:05:33
0:05:33
 0:00:30
0:00:30
 0:01:28
0:01:28
 0:01:13
0:01:13
 0:05:21
0:05:21
 0:01:14
0:01:14
 0:01:14
0:01:14
 0:23:32
0:23:32
 0:02:11
0:02:11
 0:01:28
0:01:28
 0:00:54
0:00:54
 0:16:49
0:16:49
 0:01:13
0:01:13
 0:07:04
0:07:04