filmov
tv
Single Cell RNA-Seq: full workflow in R [ public data to classified UMAP in 30 mins]
Показать описание
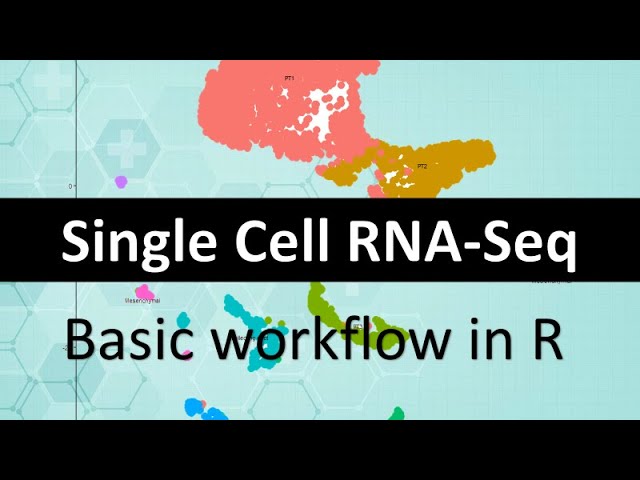
Here is a full, basic single cell RNA-Seq workflow in R, starting with some aligned publicly available data and ending with a nice UMAP plot with classified clusters and their markers banked down.
Single cell RNA-Seq has captured a lot of attention and imaginations (and publications !) in bioinformatics these last few years. Many starting Phd’s (including me ) needed to suddenly pivot and learn the basics of the field. Theres a steep learning curve, and the most valuable learning comes from iterating over your analysis, and exploring the functions and the stats behind them, and comparing your analysis to others, reading their methods etc.
Getting those first few plots up can seem like quite the hurdle and seeing a pretty tSNE or UMAP is a huge morale boost when youre learning so I think this is a good place to start. The “actual” start is of course the bench, and the start of the computational work is on the .fastq files as they come off the sequencer , but I found that phase even harder to learn actually, so I think its helpful to go for the higher yield, quicker wins first to build some confidence and have some data you can show your team at the lab meeting and start thinking about!
The data was generated using a droplet based workflow by 10x genomics but this can apply to any workflow.
## script here so you can follow along
## home of Seurat package, excellent vignettes
## another excellent course
## UMAP original paper
## viridis
Single cell RNA-Seq has captured a lot of attention and imaginations (and publications !) in bioinformatics these last few years. Many starting Phd’s (including me ) needed to suddenly pivot and learn the basics of the field. Theres a steep learning curve, and the most valuable learning comes from iterating over your analysis, and exploring the functions and the stats behind them, and comparing your analysis to others, reading their methods etc.
Getting those first few plots up can seem like quite the hurdle and seeing a pretty tSNE or UMAP is a huge morale boost when youre learning so I think this is a good place to start. The “actual” start is of course the bench, and the start of the computational work is on the .fastq files as they come off the sequencer , but I found that phase even harder to learn actually, so I think its helpful to go for the higher yield, quicker wins first to build some confidence and have some data you can show your team at the lab meeting and start thinking about!
The data was generated using a droplet based workflow by 10x genomics but this can apply to any workflow.
## script here so you can follow along
## home of Seurat package, excellent vignettes
## another excellent course
## UMAP original paper
## viridis
Комментарии
 0:02:57
0:02:57
 0:02:35
0:02:35
 0:24:37
0:24:37
 0:17:41
0:17:41
 0:34:35
0:34:35
 0:05:50
0:05:50
 0:02:39
0:02:39
 0:12:11
0:12:11
 0:40:32
0:40:32
 1:18:40
1:18:40
 0:03:46
0:03:46
 0:02:01
0:02:01
 2:50:44
2:50:44
 0:36:18
0:36:18
 0:12:24
0:12:24
 0:38:23
0:38:23
 0:02:58
0:02:58
 0:36:09
0:36:09
 0:54:15
0:54:15
 0:07:33
0:07:33
 0:18:26
0:18:26
 0:04:39
0:04:39
 0:24:06
0:24:06
 0:46:35
0:46:35