filmov
tv
Gene sequencing | DNA sequencing | Sanger method | Automated DNA sequencing

Показать описание
Aslam o alikum
I am Hassam ur Rahman and I am teaching Fsc biology since 2014 .I am always trying to improve myself and provide best lecture to students.i am taking content for the lecture from authentic and relevant sources but human errors are possible . you are requested to please highlight the mistakes.My lectures are equally reliable for Fsc and mdcat students
#visiblescience #mdcatbiology #alevelbiology #neetbiology #fscbiology #genesequencing #dnasequencing
This lecture is about
Gene Sequencing
In the late 1970s, methods were developed that allowed the nucleotide sequence of
any purified DNA fragement to be determined simply and quickly. The main principle
of these methods is :
1. To generate pieces of DNA of different sizes all starting from the same point and
ending at different points.
2. Separation of these different pieces of DNA on agarose gel.
3. Reading of sequence from the gel.
For generation of different sized DNA fragments, two methods arc generally used. One
is Sanger’s,method in which dideoxyribonucleoside triphosphates arc used to terminate
DNA synthesis at different sites. The other method is known as Maxam-Giibcrt method
in which DNA threads are chemically cut into pieces of different sizes
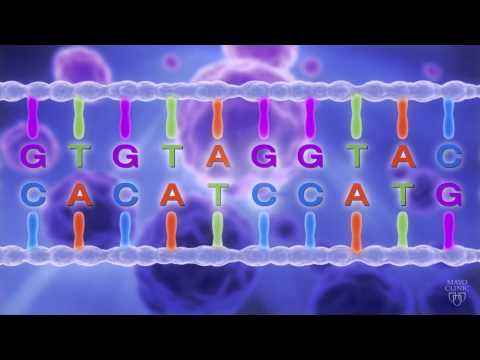
Fig 23.8 shows typical gel obtained after dideoxy method. The volume of DNA sequence
information is now so large that powerful computers must be used to store and analyze
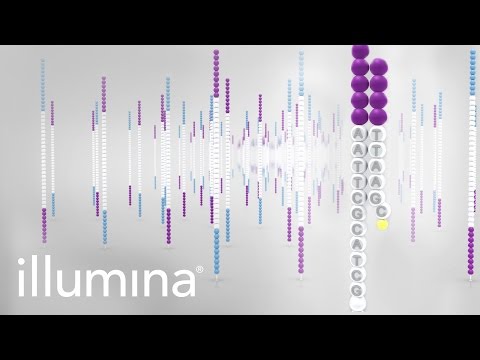
it. DNA sequence is now completely automated, robotic devices mix the reagents and
then load, run and read the order of the nucleotide bases from the gel. This is facilitated
by using chain terminating nucleotides that are each labelled with a different colored
fluorescent dye; in this case, all four synthesis reactions can be performed in the same
tube, and the products can be separated in a single lane of a gel. A detector positioned
near the bottom of the gel reads and records the color of fluorescent label on each band
as it passes through a laser beam. A computer then reads and stores this nucleotide
sequence.
Owing to the automation of DNA sequencing, the genomes of many organisms have
been sequenced. These include plant chloroplasts and animal mitochondria, large
number of bacteria, many of the yeasts, a nematode worm. Drosophila, the model plant
Arabidopsis, the mouse and human. Researchers have also deduced the complete DNA
sequence of a variety of human pathogens
I am Hassam ur Rahman and I am teaching Fsc biology since 2014 .I am always trying to improve myself and provide best lecture to students.i am taking content for the lecture from authentic and relevant sources but human errors are possible . you are requested to please highlight the mistakes.My lectures are equally reliable for Fsc and mdcat students
#visiblescience #mdcatbiology #alevelbiology #neetbiology #fscbiology #genesequencing #dnasequencing
This lecture is about
Gene Sequencing
In the late 1970s, methods were developed that allowed the nucleotide sequence of
any purified DNA fragement to be determined simply and quickly. The main principle
of these methods is :
1. To generate pieces of DNA of different sizes all starting from the same point and
ending at different points.
2. Separation of these different pieces of DNA on agarose gel.
3. Reading of sequence from the gel.
For generation of different sized DNA fragments, two methods arc generally used. One
is Sanger’s,method in which dideoxyribonucleoside triphosphates arc used to terminate
DNA synthesis at different sites. The other method is known as Maxam-Giibcrt method
in which DNA threads are chemically cut into pieces of different sizes
Fig 23.8 shows typical gel obtained after dideoxy method. The volume of DNA sequence
information is now so large that powerful computers must be used to store and analyze
it. DNA sequence is now completely automated, robotic devices mix the reagents and
then load, run and read the order of the nucleotide bases from the gel. This is facilitated
by using chain terminating nucleotides that are each labelled with a different colored
fluorescent dye; in this case, all four synthesis reactions can be performed in the same
tube, and the products can be separated in a single lane of a gel. A detector positioned
near the bottom of the gel reads and records the color of fluorescent label on each band
as it passes through a laser beam. A computer then reads and stores this nucleotide
sequence.
Owing to the automation of DNA sequencing, the genomes of many organisms have
been sequenced. These include plant chloroplasts and animal mitochondria, large
number of bacteria, many of the yeasts, a nematode worm. Drosophila, the model plant
Arabidopsis, the mouse and human. Researchers have also deduced the complete DNA
sequence of a variety of human pathogens
Комментарии
 0:02:11
0:02:11
 0:04:55
0:04:55
 0:04:41
0:04:41
 0:01:31
0:01:31
 0:48:07
0:48:07
 0:03:28
0:03:28
 0:01:37
0:01:37
 0:05:05
0:05:05
 0:08:31
0:08:31
 0:04:34
0:04:34
 0:04:12
0:04:12
 0:31:26
0:31:26
 0:03:53
0:03:53
 0:03:28
0:03:28
 0:06:36
0:06:36
 0:09:30
0:09:30
 0:05:13
0:05:13
 0:01:10
0:01:10
 0:15:17
0:15:17
 0:03:26
0:03:26
 0:09:33
0:09:33
 0:01:40
0:01:40
 0:03:00
0:03:00
 0:01:30
0:01:30