filmov
tv
Next Generation Sequencing (ILLUMINA SEQUENCING)

Показать описание
0:00-1:07 | What is the human genome project?
1:07-1:47 | What is Next Generation Sequencing?
1:47-2:25 | How does Illumina sequencing work?
2:25-3:23 | How does Bridge Amplification work?
3:23-4:03 | How does Sequencing by synthesis work?
4:03-4:41 | How does the alignment and data analysis in illumina sequencing work?
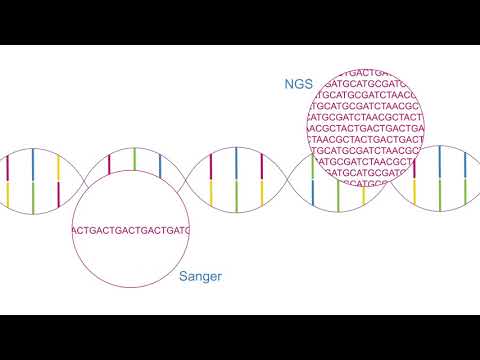
This is how many book volumes it would take to contain the entirety of a sequenced human genome. The human genome (or actually only about 90% of it) was first sequenced by the Human Genome Project back in 2003. The project was started in 1990 meaning that it took a total of 13 years to sequence this first genome. Moreover it cost a massive 3 BILLION dollars, yeah that is Billion with a B. The DNA sequencing method used to do this was Sanger sequencing, which is something I have already covered on the channel in this video!
Now, today in 2022, the current world record for sequencing the entirety of a human genome is… checks the internet… 5 HOURS AND 2 MINUTES! Wow! Even I am shocked by that! The price? 399-1000 dollars… But how can this be? Well this is thanks to something called Next-generation sequencing (NGS), also known as high-throughput sequencing. However, to achieve this particular feat an upgraded version of NGS was used. But what is Next-generation sequencing and why is it so powerful?
Next-generation sequencing (NGS), also known as high-throughput sequencing, is the catch-all term used to describe a number of different modern sequencing technologies. These technologies allow for sequencing of DNA and RNA much more quickly and cheaply compared to Sanger sequencing.
NGS technologies include:
1. Illumina (Solexa) sequencing, which we will examine closer today
2. Roche 454 sequencing which I have already covered in this video…
3. Ion Torrent sequencing
Illumina sequencing can be divided into 4 main stages, library preparation, cluster amplification, sequencing and finally alignment and data analysis.
1. During library preparation, genomic DNA is fragmented into small segments. Then specialized adaptors are attached to both end of each segment through ligation.
2. The library is then loaded into a flow cell or chip and is amplified into a clonal cluster through bridge amplification. This is done in the following manner:
1. First, the DNA segment is amplified through PCR
2. Then the original segment is washed away
3. Then the segments bends, attaching itself to the adjacent adaptors present on the chip
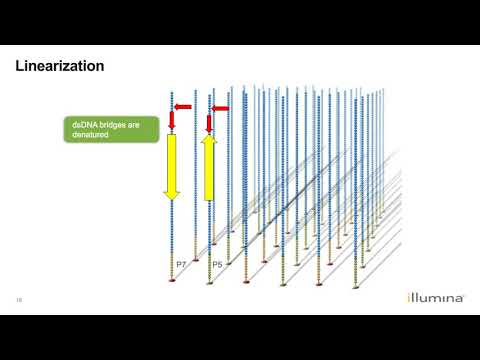
4. This “bridge is then amplified using PCR and denatured
5. In this way, bridge amplification may continue creating a cluster of clones in the process
6. Before the sequencing can start, the reverse strand is cleaved.
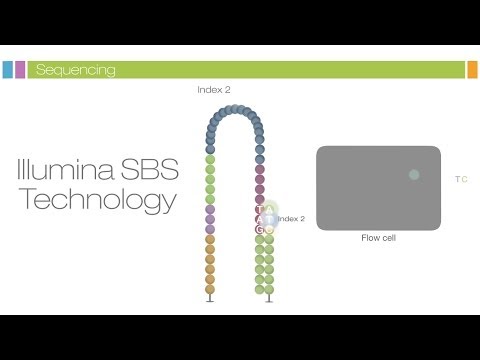
3. Now, these different clonal clusters can be sequenced through synthesis. We again examine this process in detail by only focusing on a single segment:
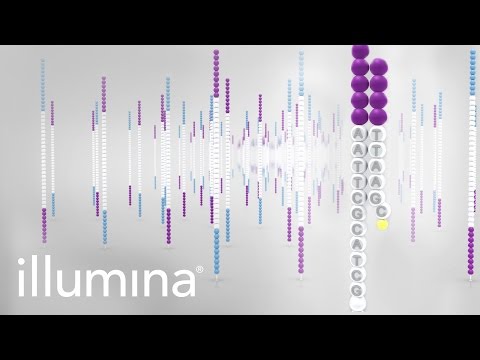
1. Sequencing reagents are added, including nucleotides which are fluorescently labeled, meaning that we can detect the nucleotide when it attaches to the segment.
2. This can be done thanks to the wavelength and intensity emitted by each of the fluorophores attached to the nucleotides when we shine a laser on them.
3. This is done simultaneously for all clonal clusters to create individual “reads”.
4. Now, we can align these generated reads to a reference genome with bioinformatics software. After alignment, any differences between the newly sequenced reads and the reference genome can be identified.
1:07-1:47 | What is Next Generation Sequencing?
1:47-2:25 | How does Illumina sequencing work?
2:25-3:23 | How does Bridge Amplification work?
3:23-4:03 | How does Sequencing by synthesis work?
4:03-4:41 | How does the alignment and data analysis in illumina sequencing work?
This is how many book volumes it would take to contain the entirety of a sequenced human genome. The human genome (or actually only about 90% of it) was first sequenced by the Human Genome Project back in 2003. The project was started in 1990 meaning that it took a total of 13 years to sequence this first genome. Moreover it cost a massive 3 BILLION dollars, yeah that is Billion with a B. The DNA sequencing method used to do this was Sanger sequencing, which is something I have already covered on the channel in this video!
Now, today in 2022, the current world record for sequencing the entirety of a human genome is… checks the internet… 5 HOURS AND 2 MINUTES! Wow! Even I am shocked by that! The price? 399-1000 dollars… But how can this be? Well this is thanks to something called Next-generation sequencing (NGS), also known as high-throughput sequencing. However, to achieve this particular feat an upgraded version of NGS was used. But what is Next-generation sequencing and why is it so powerful?
Next-generation sequencing (NGS), also known as high-throughput sequencing, is the catch-all term used to describe a number of different modern sequencing technologies. These technologies allow for sequencing of DNA and RNA much more quickly and cheaply compared to Sanger sequencing.
NGS technologies include:
1. Illumina (Solexa) sequencing, which we will examine closer today
2. Roche 454 sequencing which I have already covered in this video…
3. Ion Torrent sequencing
Illumina sequencing can be divided into 4 main stages, library preparation, cluster amplification, sequencing and finally alignment and data analysis.
1. During library preparation, genomic DNA is fragmented into small segments. Then specialized adaptors are attached to both end of each segment through ligation.
2. The library is then loaded into a flow cell or chip and is amplified into a clonal cluster through bridge amplification. This is done in the following manner:
1. First, the DNA segment is amplified through PCR
2. Then the original segment is washed away
3. Then the segments bends, attaching itself to the adjacent adaptors present on the chip
4. This “bridge is then amplified using PCR and denatured
5. In this way, bridge amplification may continue creating a cluster of clones in the process
6. Before the sequencing can start, the reverse strand is cleaved.
3. Now, these different clonal clusters can be sequenced through synthesis. We again examine this process in detail by only focusing on a single segment:
1. Sequencing reagents are added, including nucleotides which are fluorescently labeled, meaning that we can detect the nucleotide when it attaches to the segment.
2. This can be done thanks to the wavelength and intensity emitted by each of the fluorophores attached to the nucleotides when we shine a laser on them.
3. This is done simultaneously for all clonal clusters to create individual “reads”.
4. Now, we can align these generated reads to a reference genome with bioinformatics software. After alignment, any differences between the newly sequenced reads and the reference genome can be identified.
Комментарии
 0:04:44
0:04:44
 0:05:13
0:05:13
 0:41:35
0:41:35
 0:05:04
0:05:04
 0:25:05
0:25:05
 0:04:23
0:04:23
 0:31:26
0:31:26
 0:04:02
0:04:02
 0:12:29
0:12:29
 0:04:09
0:04:09
 0:03:44
0:03:44
 0:04:25
0:04:25
 0:03:40
0:03:40
 0:09:30
0:09:30
 0:04:41
0:04:41
 0:01:15
0:01:15
 0:03:21
0:03:21
 0:02:51
0:02:51
 0:51:48
0:51:48
 0:12:53
0:12:53
 0:43:32
0:43:32
 0:07:34
0:07:34
 0:04:22
0:04:22
 0:04:49
0:04:49