filmov
tv
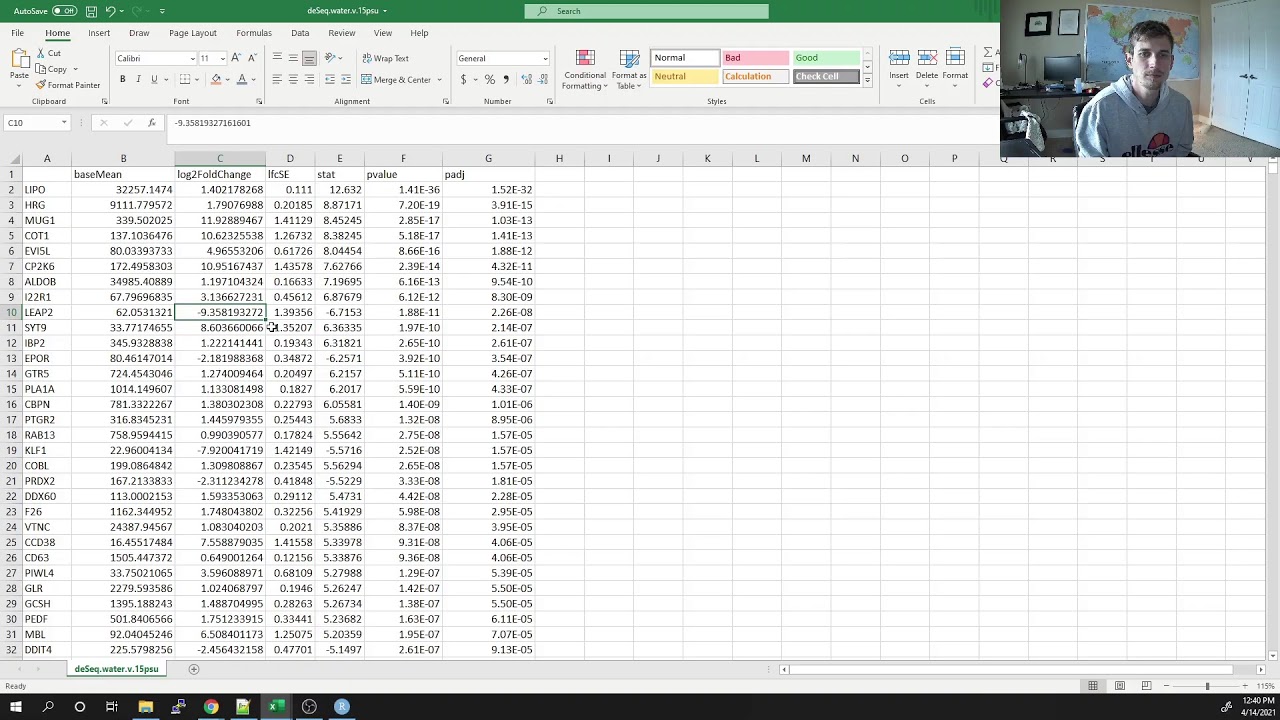
DESeq basics
Показать описание
DESeq basics
DESeq2 Basics Explained | Differential Gene Expression Analysis | Bioinformatics 101
DESeq2 workflow tutorial | Differential Gene Expression Analysis | Bioinformatics 101
deseq tutorial & visualization. how to plot dispersion estimates
How I analyze RNA Seq Gene Expression data using DESeq2
DESeq2 workflow tutorial | Differential Gene Expression Analysis | RNA Seq
RNA Seq deseq tutorial & visualization | heatmaps
Differential Gene Expression Analysis in R with DESeq2| Bioinformatics Tutorial for Beginners
Bioinformatic tools online - Differential Gene Expression analysis (DESeq2, DESeq, EdgeR)
DeSeq - Differential Gene Expression Analysis on RNA-seq data - R Tutorial
RNAseq tutorial – part 4 – Differential expression analysis with Deseq2
Multifactor Designs in DESeq2
Deseq: Multiple Comparisons
Deseq: Two way comparison
Intro to Bioinformatics - EdgeR vs DESeq2 - VDB Computational Biology
Shrinkage DESeq2
Time for DESEQ RNAseq Analysis! (John Rinn Computational Genomics Lab)
DESeq pt 1
DESeq plotting
RNA Seq deseq tutorial & visualization | PCA plot with R
RNA Seq deseq tutorial & visualization | ma plot with R
DESeq pt 2
Read Counting and Differential Gene Expression using DeSeq
RNAseq -09 - DEseq Analysis
Комментарии
 0:44:22
0:44:22
 0:25:32
0:25:32
 0:19:46
0:19:46
 0:01:58
0:01:58
 1:18:01
1:18:01
 1:19:30
1:19:30
 0:23:52
0:23:52
 0:30:36
0:30:36
 0:12:54
0:12:54
 0:08:30
0:08:30
 0:16:23
0:16:23
 0:21:58
0:21:58
 0:21:21
0:21:21
 0:31:17
0:31:17
 0:17:13
0:17:13
 0:06:23
0:06:23
 0:10:29
0:10:29
 0:51:24
0:51:24
 0:34:24
0:34:24
 0:07:39
0:07:39
 0:04:41
0:04:41
 1:02:20
1:02:20
 0:09:48
0:09:48
 0:03:27
0:03:27