filmov
tv
Visualizing gene knock-outs using RNA-Seq data in the Integrative Genomics Viewer (IGV)
Показать описание
-----
In this video step, we will do some fun and fancy visualization of RNA-Seq data using the Integrative Genomics Viewer.
IGV is a desktop tool so we'll start it up with a slightly different method than other GenomeSpace tools. Right-click the IGV icon and choose "Launch" from the context menu.
Open the downloaded JNLP file. Ignore any warnings that may pop up.
We will be using IGV to view some BAM files.
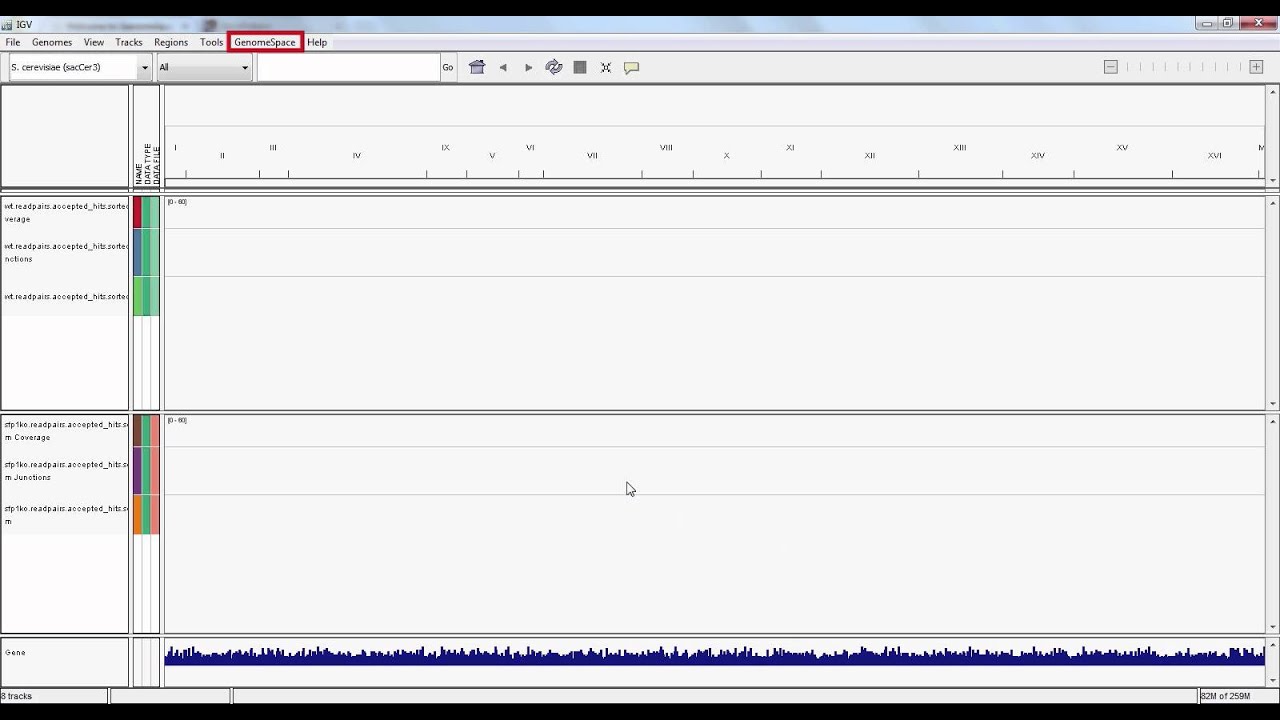
Remember to check that the genome displayed in the top left corner corresponds to your BAM file's build; in this case, "S. cerevisiae sacCer3". You can change this parameter via the drop-down menu.
We should have three (3) visualized genomic tracks.
We will next use IGV to confirm the knock-out of gene YLR403W in our mutant S. cerevisiae.
In the search bar, type "YLR403W" and hit enter. The search bar will display corresponding chromosome coordinates while loading the data which may take some time.
Notice that our wild-type track displays reads at this gene, but the mutant track shows no hits. That is, we have successfully knocked out the gene YLR403W in our mutant yeast strain.
 0:02:08
0:02:08
 0:49:56
0:49:56
 0:03:38
0:03:38
 0:02:08
0:02:08
 0:24:51
0:24:51
 0:18:34
0:18:34
 0:38:52
0:38:52
 0:01:39
0:01:39
 1:26:05
1:26:05
 0:43:12
0:43:12
 0:04:32
0:04:32
 0:05:32
0:05:32
 0:01:19
0:01:19
 0:04:12
0:04:12
 0:31:11
0:31:11
 0:34:20
0:34:20
 0:57:16
0:57:16
 0:55:11
0:55:11
 0:56:10
0:56:10
 0:55:23
0:55:23
 0:55:36
0:55:36
 0:40:59
0:40:59
 0:05:46
0:05:46
 0:00:10
0:00:10