filmov
tv
Contrastive and neighbor embedding methods for data visualisation - Dmitry Kobak
Показать описание
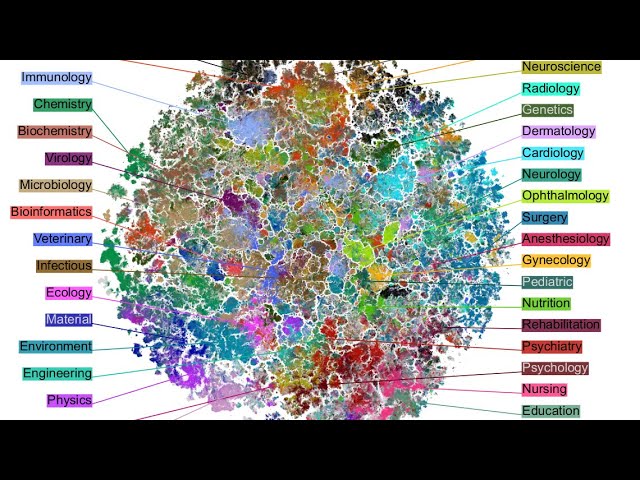
In recent years, neighbor embedding methods like t-SNE and UMAP have become widely used across several application fields, in particular in single-cell biology. They are also widely used for visualising large collections of documents and/or images used to train modern deep learning architectures such as large language models or diffusion models. Given this academic and public attention, it is very important to understand possibilities, shortcomings, and trade-offs of neighbor embedding methods. I am going to present our recent work on the attraction-repulsion spectrum of neighbor embeddings and the involved trade-offs. I am also going to explain how neighbor embeddings are related to contrastive learning, a popular framework for self-supervised learning of image data. This will lead to our recent work on contrastive visualizations of image datasets. In the second part of the talk, I will present our ongoing work on visualization of scientific literature, in particular biomedical research papers from the PubMed library.
Dmitry Kobak is a research scientist and a group leader in the Berens lab at Tübingen University, Germany. He is interested in unsupervised and self-supervised learning, in particular contrastive learning, manifold learning, and dimensionality reduction for 2D visualization of biological datasets.
Dmitry Kobak is a research scientist and a group leader in the Berens lab at Tübingen University, Germany. He is interested in unsupervised and self-supervised learning, in particular contrastive learning, manifold learning, and dimensionality reduction for 2D visualization of biological datasets.
Комментарии
 0:43:01
0:43:01
 1:39:30
1:39:30
 0:13:01
0:13:01
 1:00:01
1:00:01
 0:18:46
0:18:46
 0:41:15
0:41:15
 0:22:42
0:22:42
 0:37:00
0:37:00
 0:14:21
0:14:21
 0:18:47
0:18:47
 0:08:39
0:08:39
 0:01:44
0:01:44
 0:50:55
0:50:55
 0:18:52
0:18:52
 0:21:11
0:21:11
 0:56:53
0:56:53
 0:39:21
0:39:21
 0:20:52
0:20:52
 0:02:36
0:02:36
 1:16:30
1:16:30
 1:07:15
1:07:15
 0:16:07
0:16:07
 1:06:37
1:06:37
 1:06:37
1:06:37