filmov
tv
RNA Editing Process

Показать описание
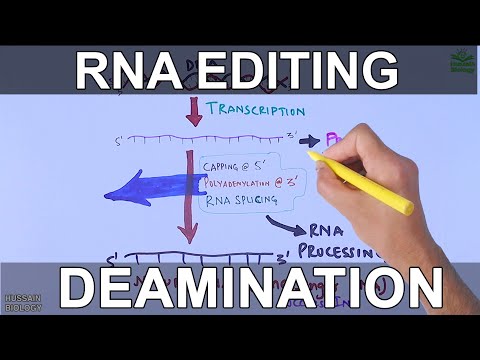
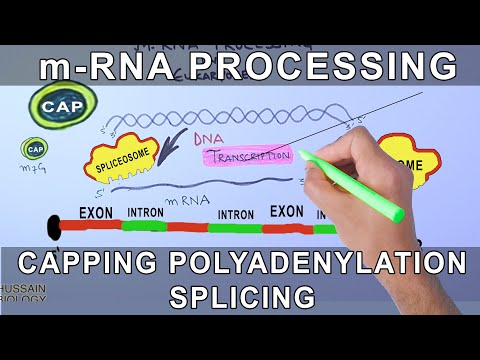
RNA editing (also RNA modification) is a molecular process through which some cells can make discrete changes to specific nucleotide sequences within an RNA molecule after it has been generated by RNA polymerase. It occurs in all living organisms and is one of the most evolutionarily conserved properties of RNAs.RNA editing may include the insertion, deletion, and base substitution of nucleotides within the RNA molecule. RNA editing is relatively rare, with common forms of RNA processing (e.g. splicing, 5'-capping, and 3'-polyadenylation) not usually considered as editing. It can affect the activity, localization as well as stability of RNAs, and has been linked with human diseases.
RNA editing has been observed in some tRNA, rRNA, mRNA, or miRNA molecules of eukaryotes and their viruses, archaea, and prokaryotes.[5] RNA editing occurs in the cell nucleus, as well as within mitochondria and plastids. In vertebrates, editing is rare and usually consists of a small number of changes to the sequence of the affected molecules. In other organisms, such as squids,extensive editing (pan-editing) can occur; in some cases the majority of nucleotides in an mRNA sequence may result from editing. More than 160 types of RNA modifications have been described so far.
RNA-editing processes show great molecular diversity, and some appear to be evolutionarily recent acquisitions that arose independently. The diversity of RNA editing phenomena includes nucleobase modifications such as cytidine (C) to uridine (U) and adenosine (A) to inosine (I) deaminations, as well as non-template nucleotide additions and insertions. RNA editing in mRNAs effectively alters the amino acid sequence of the encoded protein so that it differs from that predicted by the genomic DNA sequence.
RNA editing through the addition and deletion of uracil has been found in kinetoplasts from the mitochondria of Trypanosoma brucei.Because this may involve a large fraction of the sites in a gene, it is sometimes called "pan-editing" to distinguish it from topical editing of one or a few sites.
Pan-editing starts with the base-pairing of the unedited primary transcript with a guide RNA (gRNA), which contains complementary sequences to the regions around the insertion/deletion points. The newly formed double-stranded region is then enveloped by an editosome, a large multi-protein complex that catalyzes the editing. The editosome opens the transcript at the first mismatched nucleotide and starts inserting uridines. The inserted uridines will base-pair with the guide RNA, and insertion will continue as long as A or G is present in the guide RNA and will stop when a C or U is encountered. The inserted nucleotides cause a frameshift, and result in a translated protein that differs from its gene.
RNA editing has been observed in some tRNA, rRNA, mRNA, or miRNA molecules of eukaryotes and their viruses, archaea, and prokaryotes.[5] RNA editing occurs in the cell nucleus, as well as within mitochondria and plastids. In vertebrates, editing is rare and usually consists of a small number of changes to the sequence of the affected molecules. In other organisms, such as squids,extensive editing (pan-editing) can occur; in some cases the majority of nucleotides in an mRNA sequence may result from editing. More than 160 types of RNA modifications have been described so far.
RNA-editing processes show great molecular diversity, and some appear to be evolutionarily recent acquisitions that arose independently. The diversity of RNA editing phenomena includes nucleobase modifications such as cytidine (C) to uridine (U) and adenosine (A) to inosine (I) deaminations, as well as non-template nucleotide additions and insertions. RNA editing in mRNAs effectively alters the amino acid sequence of the encoded protein so that it differs from that predicted by the genomic DNA sequence.
RNA editing through the addition and deletion of uracil has been found in kinetoplasts from the mitochondria of Trypanosoma brucei.Because this may involve a large fraction of the sites in a gene, it is sometimes called "pan-editing" to distinguish it from topical editing of one or a few sites.
Pan-editing starts with the base-pairing of the unedited primary transcript with a guide RNA (gRNA), which contains complementary sequences to the regions around the insertion/deletion points. The newly formed double-stranded region is then enveloped by an editosome, a large multi-protein complex that catalyzes the editing. The editosome opens the transcript at the first mismatched nucleotide and starts inserting uridines. The inserted uridines will base-pair with the guide RNA, and insertion will continue as long as A or G is present in the guide RNA and will stop when a C or U is encountered. The inserted nucleotides cause a frameshift, and result in a translated protein that differs from its gene.
Комментарии
 0:03:15
0:03:15
 0:22:51
0:22:51
 0:06:39
0:06:39
 0:02:05
0:02:05
 0:01:38
0:01:38
 0:07:08
0:07:08
 0:02:05
0:02:05
 0:03:17
0:03:17
 0:09:05
0:09:05
 0:03:32
0:03:32
 0:05:45
0:05:45
 0:09:06
0:09:06
 0:10:03
0:10:03
 0:03:39
0:03:39
 0:14:34
0:14:34
 0:11:57
0:11:57
 0:02:50
0:02:50
 0:04:30
0:04:30
 0:00:47
0:00:47
 0:11:27
0:11:27
 0:20:58
0:20:58
 0:02:00
0:02:00
 0:00:09
0:00:09
 0:10:24
0:10:24