filmov
tv
Charlie Boone - The Genetic Landscape of a Cell

Показать описание
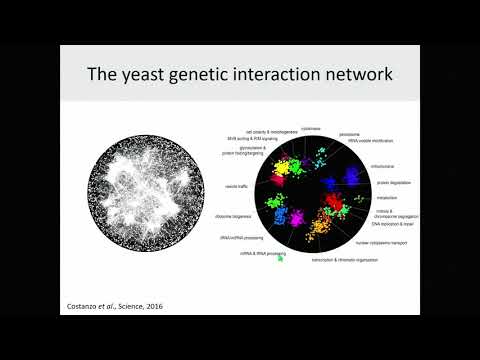
We generated a global genetic interaction network for Saccharomyces cerevisiae, testing most of the 18 million possible gene pairs for genetic interactions, identifying ~550,000 negative and ~350,000 positive genetic interactions. This comprehensive network maps genetic interactions for
essential gene pairs, highlighting essential genes as densely connected hubs. Genetic interaction profiles enabled assembly of a hierarchical model of cell function, including modules corresponding to protein complexes and pathways, biological processes, and cellular compartments. Negative interactions connected functionally related genes, mapped core bioprocesses, and identified pleiotropic genes, whereas positive interactions often mapped general regulatory connections
among gene pairs, rather than shared functionality. The global network illustrates how coherent sets of genetic interactions connect protein complex and pathway modules to map a functional wiring diagram of the cell. We are also exploring more complex interactions involving three different genes.
Like their digenic counterparts, trigenic interactions often occurred among functionally related genes and essential genes were hubs on the trigenic network. Despite their functional enrichment, trigenic interactions tended to link genes in distant bioprocesses. Nevertheless, the digenic network underlies the trigenic network because trigenic interactions often overlap a digenic interactions. Astoundingly, we estimate that the global trigenic interaction network is ~100
-
fold larger than the
global digenic network, highlighting the potential for complex genetic interactions to impact the
biology of inheritance, including the genotype to phenotype relationship.
essential gene pairs, highlighting essential genes as densely connected hubs. Genetic interaction profiles enabled assembly of a hierarchical model of cell function, including modules corresponding to protein complexes and pathways, biological processes, and cellular compartments. Negative interactions connected functionally related genes, mapped core bioprocesses, and identified pleiotropic genes, whereas positive interactions often mapped general regulatory connections
among gene pairs, rather than shared functionality. The global network illustrates how coherent sets of genetic interactions connect protein complex and pathway modules to map a functional wiring diagram of the cell. We are also exploring more complex interactions involving three different genes.
Like their digenic counterparts, trigenic interactions often occurred among functionally related genes and essential genes were hubs on the trigenic network. Despite their functional enrichment, trigenic interactions tended to link genes in distant bioprocesses. Nevertheless, the digenic network underlies the trigenic network because trigenic interactions often overlap a digenic interactions. Astoundingly, we estimate that the global trigenic interaction network is ~100
-
fold larger than the
global digenic network, highlighting the potential for complex genetic interactions to impact the
biology of inheritance, including the genotype to phenotype relationship.
 1:04:35
1:04:35
 1:31:15
1:31:15
 0:00:12
0:00:12
 0:02:20
0:02:20
 0:46:16
0:46:16
 0:00:28
0:00:28
 0:02:23
0:02:23
 0:00:30
0:00:30
 0:00:32
0:00:32
 0:00:15
0:00:15
 0:01:44
0:01:44
 0:01:06
0:01:06
 0:05:35
0:05:35
 0:33:40
0:33:40
 0:38:29
0:38:29
 0:20:24
0:20:24
 0:00:59
0:00:59
 0:36:58
0:36:58
 0:15:38
0:15:38
 0:01:00
0:01:00
 0:00:57
0:00:57
 0:00:34
0:00:34
 0:01:33
0:01:33
 0:10:26
0:10:26