filmov
tv
scRNA-seq: Dimension reduction (PCA, tSNE, UMAP)

Показать описание
We are now done with the pre-processing of the data. It’s time to talk about dimension reduction.
We won’t go through the mathematical details, but instead aim for the intuition of how dimensional reduction methods (PCA, tSNE, UMAP) work. We want to learn how to reduce dimensions and visualise our data. We also learn how to select the principal components for the clustering step.
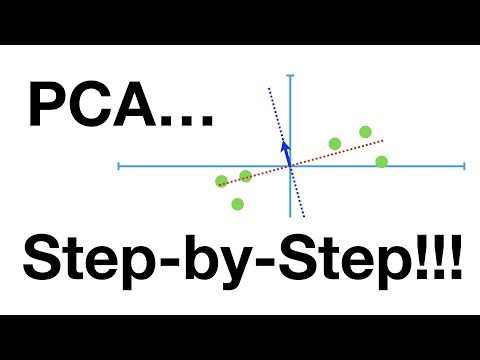
01:57 PCA
08:50 tSNE and UMAP for visualisation
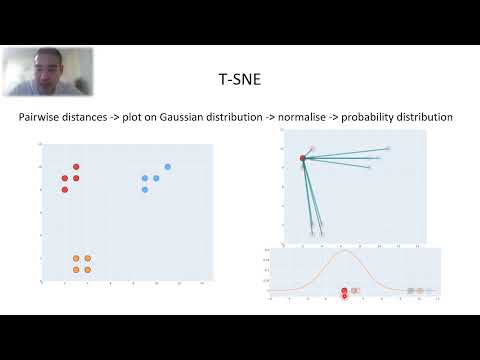
10:05 tSNE
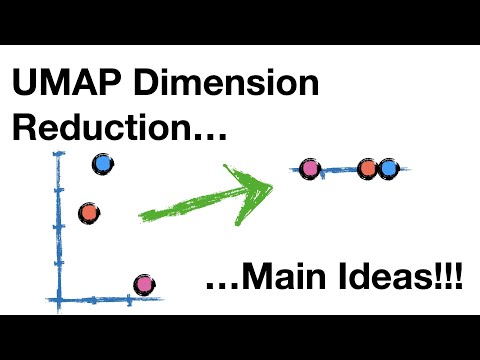
11:23 UMAP
We also recommend the excellent StatQuest videos explaining PCA, such as:
or:
Finally, the image on the slide "Other dimension reduction methods: used later for visualisation" is by Shigeo Takahashi, Issei Fujishiro, and Masato Okada, "Applying Manifold Learning to Plotting Approximate Contour Trees," IEEE Transactions on Visualization and Computer Graphics (Proceedings of IEEE Visualization / Information Visualization 2009), Vol. 15, No. 6, pp. 1185-1192, 2009.
We won’t go through the mathematical details, but instead aim for the intuition of how dimensional reduction methods (PCA, tSNE, UMAP) work. We want to learn how to reduce dimensions and visualise our data. We also learn how to select the principal components for the clustering step.
01:57 PCA
08:50 tSNE and UMAP for visualisation
10:05 tSNE
11:23 UMAP
We also recommend the excellent StatQuest videos explaining PCA, such as:
or:
Finally, the image on the slide "Other dimension reduction methods: used later for visualisation" is by Shigeo Takahashi, Issei Fujishiro, and Masato Okada, "Applying Manifold Learning to Plotting Approximate Contour Trees," IEEE Transactions on Visualization and Computer Graphics (Proceedings of IEEE Visualization / Information Visualization 2009), Vol. 15, No. 6, pp. 1185-1192, 2009.
Комментарии


















![[CS690] Lecture 16.1:](https://i.ytimg.com/vi/MPuKSzH7eAg/hqdefault.jpg)

