filmov
tv
Replication Fork Overview

Показать описание
DNA Replication (Updated)
DNA replication - 3D
Replication fork coupling
Replication Fork Overview
Cell Biology | DNA Replication 🧬
DNA Structure and Replication: Crash Course Biology #10
Semidiscontinuous DNA replication
DNA Replication - Leading Strand vs Lagging Strand & Okazaki Fragments
DNA Replication at the replication fork
Stephen P. Bell (MIT / HHMI) 1a: Chromosomal DNA Replication: The DNA Replication Fork
DNA Replication: Copying the Molecule of Life
DNA Replication
DNA Replication Fork.
Label a Replication Fork [DNA Replication]
DNA Replication in Prokaryotes | Initiation
The Replication Bubble
DNA Replication Fork
Replication of DNA class 12 | DNA Replication Helicase | Replication fork | Replication Enzymes
Bacterial DNA Replication
10.1 OriC and Replication forks
What happens at the DNA replication fork?
DNA replication in Prokaryotes
DNA Replication 3D Animation
6 Steps of DNA Replication
Комментарии
 0:08:12
0:08:12
 0:03:28
0:03:28
 0:03:29
0:03:29
 0:03:18
0:03:18
 1:07:13
1:07:13
 0:12:59
0:12:59
 0:03:04
0:03:04
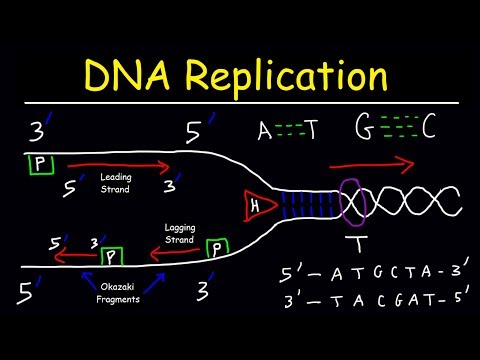 0:19:55
0:19:55
 0:09:44
0:09:44
 0:28:55
0:28:55
 0:06:16
0:06:16
 0:05:43
0:05:43
 0:02:04
0:02:04
 0:10:01
0:10:01
 0:05:53
0:05:53
 0:04:06
0:04:06
 0:01:36
0:01:36
 0:00:27
0:00:27
 0:05:40
0:05:40
 0:02:52
0:02:52
 0:00:40
0:00:40
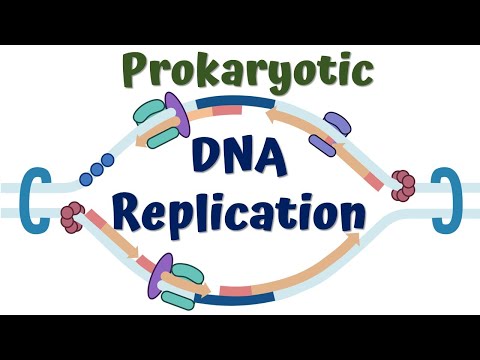 0:05:47
0:05:47
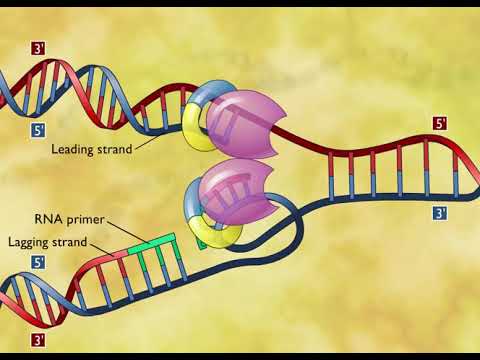 0:02:40
0:02:40
 0:17:17
0:17:17