filmov
tv
How to Efficiently Convert Row Names to Columns on Multiple Dataframes

Показать описание
Learn the best methods to convert row names into a dedicated column across multiple dataframes in R, enhancing your data analysis workflow.
---
Visit these links for original content and any more details, such as alternate solutions, latest updates/developments on topic, comments, revision history etc. For example, the original title of the Question was: How to convert row names to columns on a large number of dataframes
If anything seems off to you, please feel free to write me at vlogize [AT] gmail [DOT] com.
---
How to Efficiently Convert Row Names to Columns on Multiple Dataframes
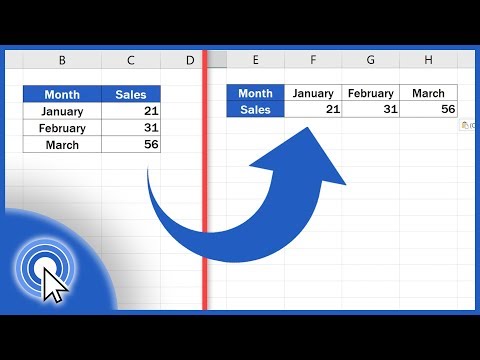
When working with large datasets in R, especially in different dataframes, you may come across a common task: converting row names into a separate column. This is particularly useful when your dataframes share identical column names but contain different row names. If you have, for instance, 100 dataframes and need to transform the row names into a column called "gene", you might be wondering how to accomplish this efficiently.
In this guide, we'll walk you through the best methods to convert row names to columns on multiple dataframes. By the end, you will have a clear understanding of the required steps and the R code necessary to make this happen seamlessly.
The Problem: Multiple Dataframes with Different Row Names
When handling numerous dataframes, it’s common to face scenarios where the row names convey valuable information but are often overlooked in analysis. Here’s the challenge broken down:
Identical Column Names: Each dataframe shares the same column structure.
Different Row Names: Each dataframe has distinct row names that need to be converted into a manageable format.
Multiple Dataframes: Performing these conversions manually on 100 dataframes is not practical.
The Solution: Using tibble::rownames_to_column
Step 1: Create a List of Dataframes
Start by gathering all your dataframes into a list. This can be done using the mget function, which allows you to collect multiple dataframe objects into a single list.
[[See Video to Reveal this Text or Code Snippet]]
Alternatively, if you want to specify each dataframe individually, you can use the dplyr::lst function:
[[See Video to Reveal this Text or Code Snippet]]
Step 2: Apply the Conversion Function
Next, apply the tibble::rownames_to_column function to each dataframe in the list. This function will take the row names from each dataframe and convert them into a new column, which we will name "gene". Use the lapply function for this task:
[[See Video to Reveal this Text or Code Snippet]]
Step 3: Reflect Changes Back to the Environment
If you want the changes made to be reflected across your original dataframes in the global environment, you can use list2env:
[[See Video to Reveal this Text or Code Snippet]]
Conclusion
By following these steps, you can efficiently convert row names into a new column called "gene" across multiple dataframes without the hassle of manually editing each one. This approach not only saves time but also streamlines your data analysis process, allowing you to focus on drawing insights rather than data wrangling.
Benefits of This Method:
Efficiency: Handle numerous dataframes in a fraction of the time.
Consistency: Ensure that all dataframes contain the "gene" column for seamless integration in analyses.
Ease of Use: Simple R functions make the process straightforward and user-friendly.
With this guide, you now have the tools and knowledge to manipulate your dataframes effectively. If you have any further questions or need assistance with R coding, don’t hesitate to ask!
---
Visit these links for original content and any more details, such as alternate solutions, latest updates/developments on topic, comments, revision history etc. For example, the original title of the Question was: How to convert row names to columns on a large number of dataframes
If anything seems off to you, please feel free to write me at vlogize [AT] gmail [DOT] com.
---
How to Efficiently Convert Row Names to Columns on Multiple Dataframes
When working with large datasets in R, especially in different dataframes, you may come across a common task: converting row names into a separate column. This is particularly useful when your dataframes share identical column names but contain different row names. If you have, for instance, 100 dataframes and need to transform the row names into a column called "gene", you might be wondering how to accomplish this efficiently.
In this guide, we'll walk you through the best methods to convert row names to columns on multiple dataframes. By the end, you will have a clear understanding of the required steps and the R code necessary to make this happen seamlessly.
The Problem: Multiple Dataframes with Different Row Names
When handling numerous dataframes, it’s common to face scenarios where the row names convey valuable information but are often overlooked in analysis. Here’s the challenge broken down:
Identical Column Names: Each dataframe shares the same column structure.
Different Row Names: Each dataframe has distinct row names that need to be converted into a manageable format.
Multiple Dataframes: Performing these conversions manually on 100 dataframes is not practical.
The Solution: Using tibble::rownames_to_column
Step 1: Create a List of Dataframes
Start by gathering all your dataframes into a list. This can be done using the mget function, which allows you to collect multiple dataframe objects into a single list.
[[See Video to Reveal this Text or Code Snippet]]
Alternatively, if you want to specify each dataframe individually, you can use the dplyr::lst function:
[[See Video to Reveal this Text or Code Snippet]]
Step 2: Apply the Conversion Function
Next, apply the tibble::rownames_to_column function to each dataframe in the list. This function will take the row names from each dataframe and convert them into a new column, which we will name "gene". Use the lapply function for this task:
[[See Video to Reveal this Text or Code Snippet]]
Step 3: Reflect Changes Back to the Environment
If you want the changes made to be reflected across your original dataframes in the global environment, you can use list2env:
[[See Video to Reveal this Text or Code Snippet]]
Conclusion
By following these steps, you can efficiently convert row names into a new column called "gene" across multiple dataframes without the hassle of manually editing each one. This approach not only saves time but also streamlines your data analysis process, allowing you to focus on drawing insights rather than data wrangling.
Benefits of This Method:
Efficiency: Handle numerous dataframes in a fraction of the time.
Consistency: Ensure that all dataframes contain the "gene" column for seamless integration in analyses.
Ease of Use: Simple R functions make the process straightforward and user-friendly.
With this guide, you now have the tools and knowledge to manipulate your dataframes effectively. If you have any further questions or need assistance with R coding, don’t hesitate to ask!
 0:01:39
0:01:39
 0:05:00
0:05:00
 0:01:47
0:01:47
 0:04:51
0:04:51
 0:00:16
0:00:16
 0:00:41
0:00:41
 0:00:35
0:00:35
 0:01:55
0:01:55
 0:48:24
0:48:24
 0:07:16
0:07:16
 0:01:29
0:01:29
 0:00:10
0:00:10
 0:00:15
0:00:15
 0:00:31
0:00:31
 0:01:56
0:01:56
 0:00:19
0:00:19
 0:00:56
0:00:56
 0:02:04
0:02:04
 0:01:00
0:01:00
 0:01:07
0:01:07
 0:00:45
0:00:45
 0:00:40
0:00:40
 0:00:32
0:00:32
 0:00:31
0:00:31