filmov
tv
Building a network from a gene list using the GeneMANIA module in Cytoscape 3
Показать описание
-----
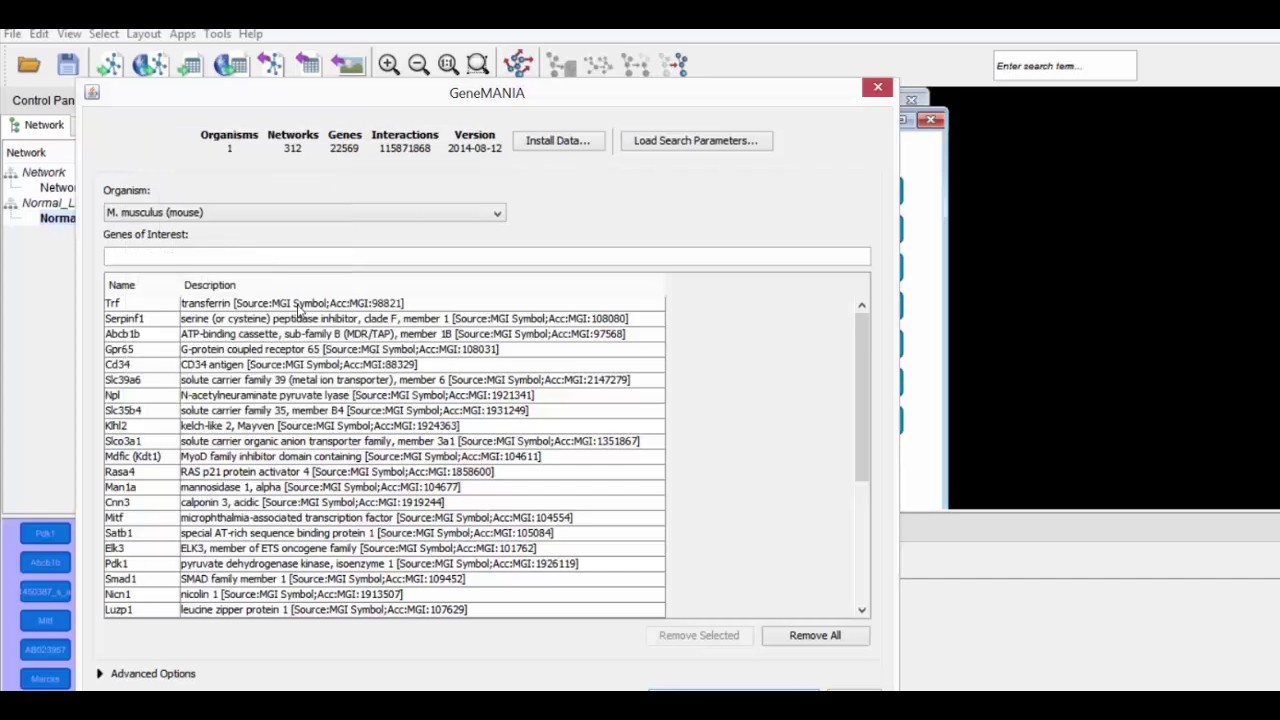
We will now use the GeneMania plug-in to find the network of interacting proteins associated with our gene list.
Make sure you have already loaded a network of gene nodes into Cytoscape 3. If this is your first time running GeneMania, directions for installation and downloading a dataset can be found in Step 7 on the recipe webpage.
Under the table panel towards the bottom, select all the node ID names by clicking the first name, then pressing shift while scrolling downward. Copy these names via control+C for later use.
Next, navigate to "Apps", "GeneMania", then "Search", in the top toolbar.
In the new window ensure that the organism selected matches the one from which you obtained your gene list. In this case, "M. musculus". Then, paste the genes of interest by pressing CTRL+V into the highlighted box. Click OK to close the window listing genes that are not recognized.
Next, click "Start" to identify connections between genes and generate the network.
Clicking a network node will highlight the corresponding gene in the "Genes" tab and provide information about that gene.
Clicking a row in the "Functions" tab will highlight all the gene nodes associated with that function.
Feel free to make use of this and any other functionalities within the Cytoscape 3 visualizer.
Комментарии
 1:00:45
1:00:45
 0:11:47
0:11:47
 0:00:21
0:00:21
 0:17:05
0:17:05
 0:08:51
0:08:51
 0:12:47
0:12:47
 0:31:28
0:31:28
 0:00:41
0:00:41
 0:00:54
0:00:54
 0:10:42
0:10:42
 0:31:27
0:31:27
 0:54:51
0:54:51
 0:02:10
0:02:10
 0:14:15
0:14:15
 0:21:45
0:21:45
 0:00:41
0:00:41
 0:02:12
0:02:12
 0:00:21
0:00:21
 0:00:50
0:00:50
 0:02:21
0:02:21
 0:17:12
0:17:12
 0:26:10
0:26:10
 0:07:50
0:07:50
 0:05:47
0:05:47