filmov
tv
Understanding SAM/BAM file specifications

Показать описание
The sequencing Alignment Mapping (SAM) file was designed to store the mapping data of a sample against a reference genome in a way that allow for easy downstream analysis.
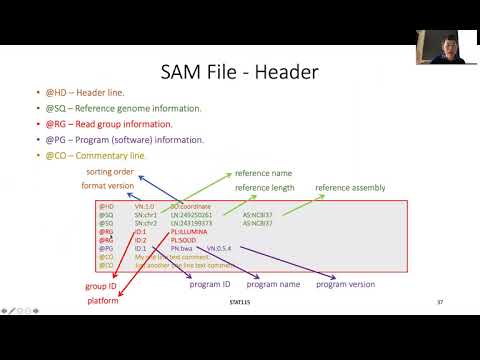
In this video, I hope to go through the basic SAM file specifications based on the original documentation from Samtools. SAM file mostly consists of two sections, a header section that records information on the reference genome and experiential details, and an alignment section that records the individual nucleotide sequence difference between the samples and the reference.
Link to SAM specifications
Link to Slides
Link to sample SAM file
How to read Cigar String
In this video, I hope to go through the basic SAM file specifications based on the original documentation from Samtools. SAM file mostly consists of two sections, a header section that records information on the reference genome and experiential details, and an alignment section that records the individual nucleotide sequence difference between the samples and the reference.
Link to SAM specifications
Link to Slides
Link to sample SAM file
How to read Cigar String
Understanding SAM/BAM file specifications
Understanding Bioinformatics File Formats: SAM/BAM
SAM flags explained | Understanding SAM flags in a SAM/BAM file | Bioinformatics 101
FASTQ, BAM, and VCF file formats easily explained - A must watch if you have had a DNA test
Samtools Tutorial | Getting the Mapping Statistics of a BAM or SAM file
Genomic file formats: BAM / SAM file format
STAT115 Chapter 3.6 SAM and BAM files
5 genomics file formats you must know
Difference between SAM and BAM
IGV (Integrative Genome Viewer) Software Tutorial | Howto visualize bam files | Episode 1
How to read Cigar Strings in SAM file
Samtools tutorial - index a bam file | samtools tutorial
SAM/BAM File Formats Session 3.3
File format system in Biological data | FASTA | FASTQ | SAM/BAM | VCF | GTF | GFF | Tutorial 7
samtools tutorial - convert sam to bam | samtools view
Tutorial-5: What are SAM/BAM/CRAM files?
FIltering bam files with samtools | remove reads from bam files episode 1
Genomics: SAM/BAM file formats
Alignment and Visualization
How to calculate depth of sequencing in SAM/BAM file?
B4B: Module 1 - NGS Data Format - BAM
Understanding File Formats in Bioinformatics: VCF and gVCF
RNA-seq Analysis 2023 | 02.2: SAM/BAM/BED file formats
SAM/BAM files and RSEM output
Комментарии
 0:18:20
0:18:20
 0:07:07
0:07:07
 0:09:27
0:09:27
 0:06:53
0:06:53
 0:02:19
0:02:19
 0:09:22
0:09:22
 0:09:16
0:09:16
 0:19:10
0:19:10
 0:00:15
0:00:15
 0:10:11
0:10:11
 0:06:51
0:06:51
 0:01:18
0:01:18
 0:21:09
0:21:09
 0:07:11
0:07:11
 0:01:33
0:01:33
 0:10:06
0:10:06
 0:02:38
0:02:38
 0:22:51
0:22:51
 0:34:14
0:34:14
 0:13:25
0:13:25
 0:35:53
0:35:53
 0:25:40
0:25:40
 0:22:12
0:22:12
 0:31:34
0:31:34