filmov
tv
How to perform phylogenetically independent contrasts

Показать описание
This tutorial shows you how to perform phylogenetically independent contrasts for a group of species, and also provides some background information about the approach and why it is used.
This tutorial uses Rstudio under Ubuntu Linux version 20.04.
This tutorial assumes you already have R and Rstudio installed. See my tutorial for installing those software:
This tutorial assumes you already have the R package "caper" installed.
This tutorial assumes you are using Ubuntu Linux version 20.04 as your operating system. If you do not have a computer running Ubuntu, fear not. You can install Ubuntu as a virtual machine within your current operating system. It is all free.
The instructions for Windows computers are here:
The instructions for Mac computers are here (from 0:00 to 5:37):
FYI:
I have made a Linux virtual machine that is pre-configured with population genetics and ecological niche modeling programs. I have made videos explaining how to hook up this virtual machine within your Mac or Windows computer, so that you can start using the pre-configured software without having to install any of them (and even without having to install Ubuntu Linux!).
How to perform phylogenetically independent contrasts
EvoBioCC Tree Thinking Clip 1 - Phylogenetic Comparative Methods
Phylogenetic comparative methods 2: independent contrasts
Clint Explains Phylogenetics - There are a million wrong ways to read a phylogenetic tree
Phylogenetic Tree Basics
Phylogenetic comparative methods 3: independent contrasts example
Phylogenetic comparative methods
History of Phylogenetic Comparative Methods (PCMs)
Phylogeny: How We're All Related: Crash Course Biology #17
Phylogenetic comparative methods
Cladistics Part 1: Constructing Cladograms
Phylogenetic comparative methods | Wikipedia audio article
AFB MycoAsia Talk “Phylogenetic Analysis for Beginners”
CSB1021 — Quick tour of phylogenetic regression
Phyloseminar #74: Josef Uyeda (Virginia Tech)
Comparative method and phylogenies
Phylogenetic tree quiz
Phylogenetic comparative methods 1: the problem
How to understand phylogenetic tree's patterns
Comparative genomics data in Ensembl, 1: Homology and gene trees
Studying adaptation using the comparative method.
Introduction to HyPhy: Hypothesis testing using Phylogenies
Using Molecular Clocks to build Phylogenetic Trees and Phylogenetic Species Concept
Virtual Phylogenetic Tree Lab
Комментарии
 0:19:55
0:19:55
 0:07:41
0:07:41
 0:12:27
0:12:27
 0:07:45
0:07:45
 0:03:34
0:03:34
 0:13:04
0:13:04
 0:13:26
0:13:26
 0:23:56
0:23:56
 0:13:51
0:13:51
 0:07:35
0:07:35
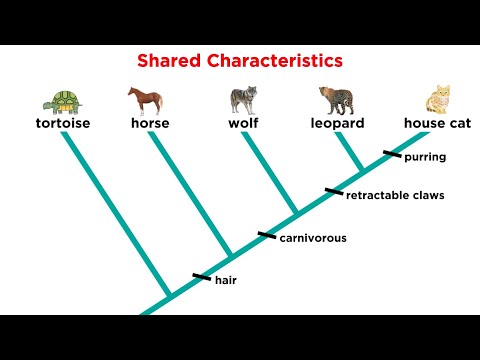 0:10:12
0:10:12
 0:10:00
0:10:00
 1:08:59
1:08:59
 0:27:44
0:27:44
 1:09:55
1:09:55
 0:59:45
0:59:45
 0:00:42
0:00:42
 0:11:29
0:11:29
 0:00:36
0:00:36
 0:18:34
0:18:34
 0:13:58
0:13:58
 0:54:35
0:54:35
 0:19:14
0:19:14
 0:08:14
0:08:14