filmov
tv
Intro to Molecular Dynamics: Coding MD From Scratch

Показать описание
This is a brief introduction to how MD simulations work: essentially numerically solving Newton’s equations for a bunch of interacting objects. It is a bit different from my usual videos because it is just something I was screwing around with for fun and thought it might be interesting to post. If you notice any issues with the code or the math please let me know. As I said, I was just screwing around so it’s not as extensively checked as publication-quality research.
Correction: When running the water simulations, there was an error in the epsilon values I was using. The videos I included are not too far off but I thought I should mention it.
Correction: As pointed out by MCMC271, simply rescaling the velocities like I did does not produce the correct probability distributions for the NVT ensemble and is thus not a good way to keep the temperature constant. I should have stated this in the video and pointed the viewer to better thermostats.
References:
(Still not sure if to include the code so it may or may not be public)
In case I do not get around to making a video on using available MD software, I recommend looking into LAMMPS, NAMD or GROMACS:
LAMMPS:
I particularly recommend the .pdf files numbered SoR 01, 05 and 11
NAMD:
GROMACS:
If you feel like paying for software, you should also check out Materials Studio and/or COMSOL.
Chapters:
0:00 Hello
0:39 Newton's equations
1:21 Code
2:30 Visualization (matplotlib)
3:10 Boundary conditions (periodic)
4:35 BCs (reflecting)
5:55 Visualization (OVITO)
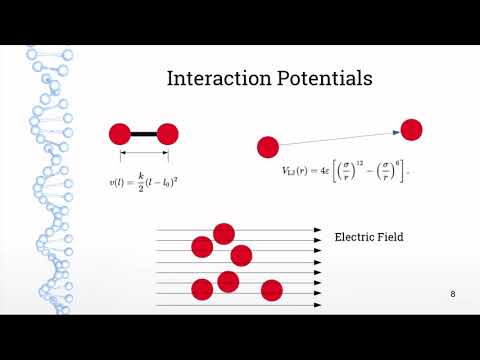
7:45 Lennard-Jones interactions
13:14 Periodic BC interaction discussion
14:48 Particle types
15:12 Microcanonical (NVE) ensemble
16:33 Canonical ensemble (fixing T)
19:26 Bond potentials
22:17 Bond angles
26:32 Dihedral angles
27:11 Electrostatics
28:28 Combining potentials
29:33 Polymers
30:08 Potential cutoff
30:53 Gravity
31:28 Summary
Correction: When running the water simulations, there was an error in the epsilon values I was using. The videos I included are not too far off but I thought I should mention it.
Correction: As pointed out by MCMC271, simply rescaling the velocities like I did does not produce the correct probability distributions for the NVT ensemble and is thus not a good way to keep the temperature constant. I should have stated this in the video and pointed the viewer to better thermostats.
References:
(Still not sure if to include the code so it may or may not be public)
In case I do not get around to making a video on using available MD software, I recommend looking into LAMMPS, NAMD or GROMACS:
LAMMPS:
I particularly recommend the .pdf files numbered SoR 01, 05 and 11
NAMD:
GROMACS:
If you feel like paying for software, you should also check out Materials Studio and/or COMSOL.
Chapters:
0:00 Hello
0:39 Newton's equations
1:21 Code
2:30 Visualization (matplotlib)
3:10 Boundary conditions (periodic)
4:35 BCs (reflecting)
5:55 Visualization (OVITO)
7:45 Lennard-Jones interactions
13:14 Periodic BC interaction discussion
14:48 Particle types
15:12 Microcanonical (NVE) ensemble
16:33 Canonical ensemble (fixing T)
19:26 Bond potentials
22:17 Bond angles
26:32 Dihedral angles
27:11 Electrostatics
28:28 Combining potentials
29:33 Polymers
30:08 Potential cutoff
30:53 Gravity
31:28 Summary
Комментарии
 0:33:25
0:33:25
 0:04:36
0:04:36
 0:18:59
0:18:59
 0:14:39
0:14:39
 0:05:43
0:05:43
 0:04:12
0:04:12
 0:03:31
0:03:31
 0:37:59
0:37:59
 0:31:30
0:31:30
 0:34:21
0:34:21
 0:00:22
0:00:22
 0:12:05
0:12:05
 0:29:07
0:29:07
 1:30:15
1:30:15
 1:55:45
1:55:45
 2:12:46
2:12:46
 0:09:45
0:09:45
 0:00:32
0:00:32
 0:03:53
0:03:53
 0:33:00
0:33:00
 0:14:00
0:14:00
 0:00:54
0:00:54
 0:07:21
0:07:21
 0:01:42
0:01:42