filmov
tv
NAMD and VMD Performance on ARM GPU Platforms, AHUG SC'20
Показать описание
Presenter: John Stone, University of Illinois
Abstract: This talk will briefly summarize and provide a updates on the current state
of GPU-accelerated ARM platform support in the NAMD parallel molecular dynamics
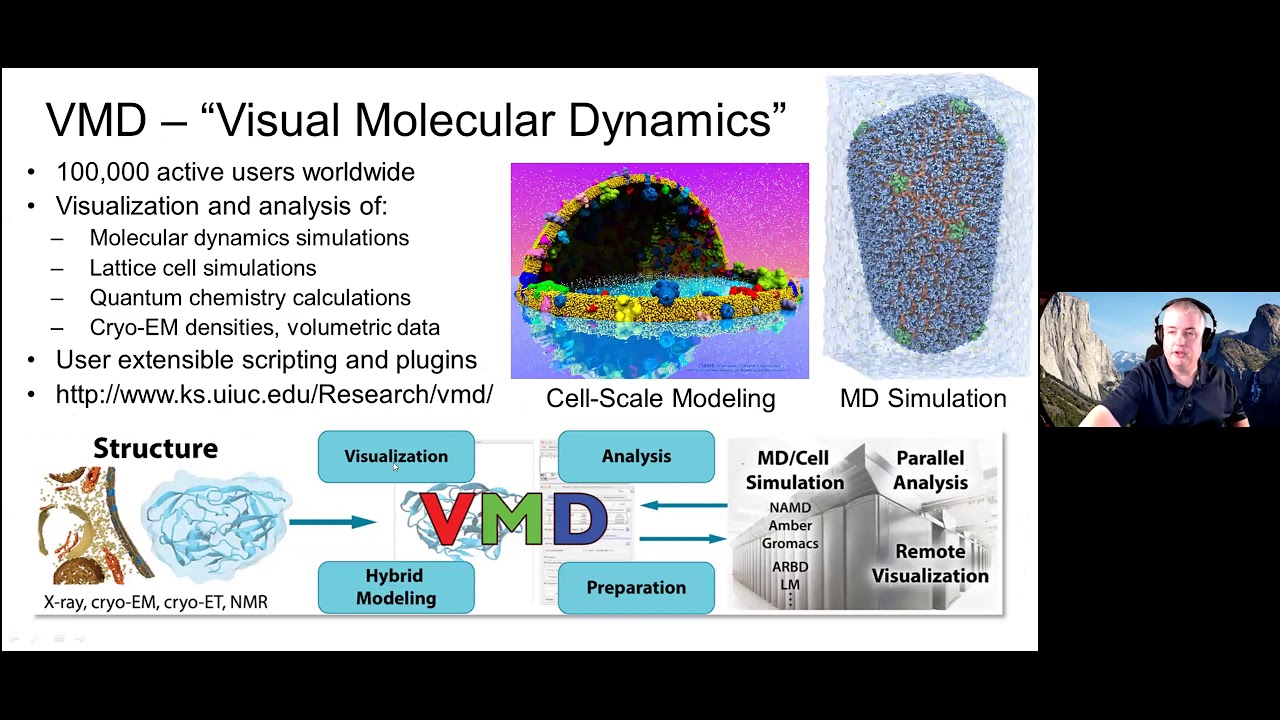
engine, and VMD, a high-performance molecular modeling environment for preparing, visualizing, and analyzing biomolecular simulations. Using the "Wombat" ARM64 cluster at Oak Ridge National Laboratory, we have recently had the opportunity to benchmark current versions of NAMD and VMD for representative molecular modeling tasks, with particular emphasis on compute heavy-operations that benefit from CUDA GPU-accelerated kernels and heterogeneous computing techniques. We highlight results that demonstrate areas where performance on the Wombat cluster is most comparable to that achieved on high-end Intel x86- and IBM POWER9-based compute nodes,
and areas where the individual strengths and weaknesses of the different platforms contribute to particular performance advantages or differences. We identify cases where our current ARM64 developments will benefit from completion of in-progress development of ARM64-specific vectorized
kernels. Finally, we present observations from very early experiences with the ARM scalable vector extensions (SVE), and vector length agnostic programming approaches, as compared with traditional vectorization on fixed-length SIMD hardware architectures.
Abstract: This talk will briefly summarize and provide a updates on the current state
of GPU-accelerated ARM platform support in the NAMD parallel molecular dynamics
engine, and VMD, a high-performance molecular modeling environment for preparing, visualizing, and analyzing biomolecular simulations. Using the "Wombat" ARM64 cluster at Oak Ridge National Laboratory, we have recently had the opportunity to benchmark current versions of NAMD and VMD for representative molecular modeling tasks, with particular emphasis on compute heavy-operations that benefit from CUDA GPU-accelerated kernels and heterogeneous computing techniques. We highlight results that demonstrate areas where performance on the Wombat cluster is most comparable to that achieved on high-end Intel x86- and IBM POWER9-based compute nodes,
and areas where the individual strengths and weaknesses of the different platforms contribute to particular performance advantages or differences. We identify cases where our current ARM64 developments will benefit from completion of in-progress development of ARM64-specific vectorized
kernels. Finally, we present observations from very early experiences with the ARM scalable vector extensions (SVE), and vector length agnostic programming approaches, as compared with traditional vectorization on fixed-length SIMD hardware architectures.
 0:17:11
0:17:11
 0:02:38
0:02:38
 0:27:08
0:27:08
 1:50:04
1:50:04
 0:20:35
0:20:35
 0:18:16
0:18:16
 0:58:19
0:58:19
 0:50:37
0:50:37
 1:01:05
1:01:05
 0:00:45
0:00:45
 0:52:29
0:52:29
 0:34:58
0:34:58
 1:20:29
1:20:29
 0:21:33
0:21:33
 0:47:29
0:47:29
 0:02:07
0:02:07
 0:01:14
0:01:14
 0:02:06
0:02:06
 0:06:37
0:06:37
 0:24:29
0:24:29
 0:25:22
0:25:22
 0:01:51
0:01:51
 0:17:34
0:17:34
 0:28:58
0:28:58