filmov
tv
Molecular dynamic simulation for protein & ligand-protein complex: web tool free easy, only click
Показать описание
In this video, I would like to show you how to perform molecular dynamic simulation for protein and ligand-protein complex: web tool free.
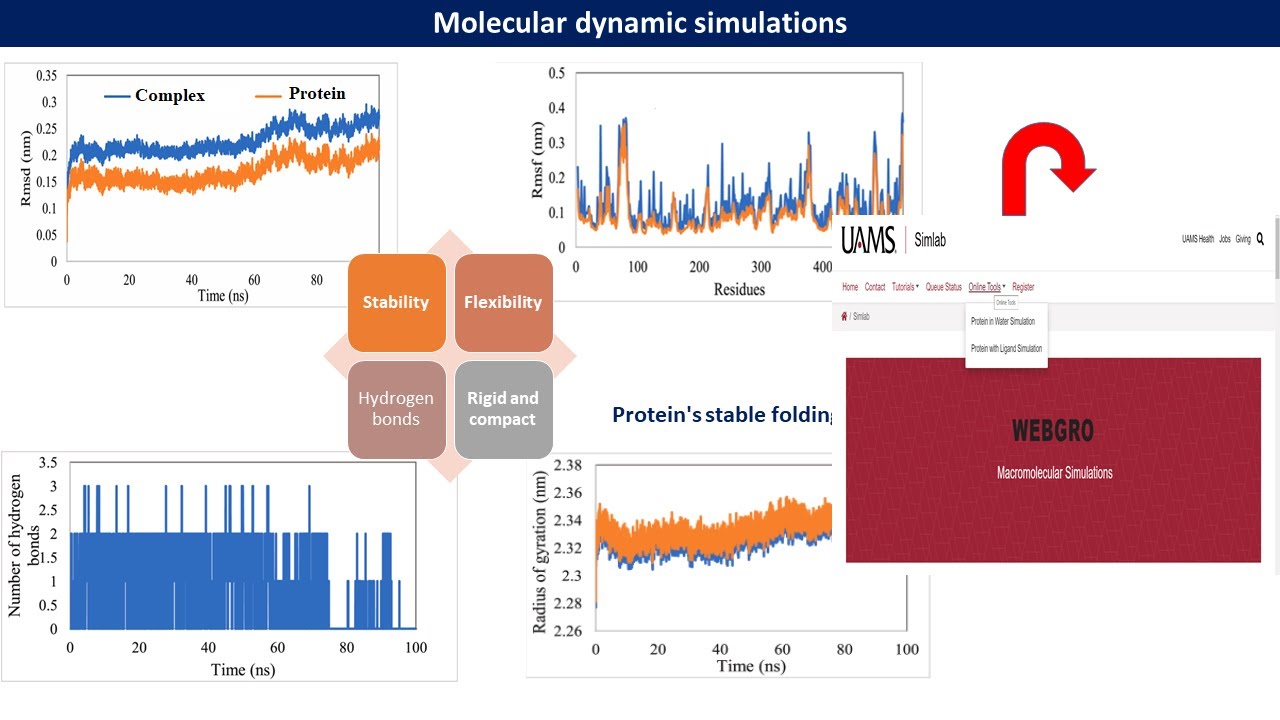
Simlab is a research initiative for creating web tools for various computational biology research including macromolecular simulations. The aim of WebGro is to make user friendly computational biology tools available for all types of researchers. WebGro is a fully automated online tool for performing molecular dynamics simulation of macromolecules (proteins) alone or in complex with ligands (small molecules).
WebGro uses the GROMACS simulation package for performing fully solvated molecular dynamics simulations. Users submit only their protein file (with .pdb extension), and WebGro will perform simulation as well as trajectory analysis. Parameters in the submission page are set to default values based on published work related to GROMACS simulations. Users are advised to refer to related papers and tutorials for in-depth understanding of each parameter.
Link:
Simlab
VMD:
Videomach
Article Related:
Simlab is a research initiative for creating web tools for various computational biology research including macromolecular simulations. The aim of WebGro is to make user friendly computational biology tools available for all types of researchers. WebGro is a fully automated online tool for performing molecular dynamics simulation of macromolecules (proteins) alone or in complex with ligands (small molecules).
WebGro uses the GROMACS simulation package for performing fully solvated molecular dynamics simulations. Users submit only their protein file (with .pdb extension), and WebGro will perform simulation as well as trajectory analysis. Parameters in the submission page are set to default values based on published work related to GROMACS simulations. Users are advised to refer to related papers and tutorials for in-depth understanding of each parameter.
Link:
Simlab
VMD:
Videomach
Article Related:
Комментарии
 0:24:29
0:24:29
 0:00:56
0:00:56
 0:00:15
0:00:15
 0:00:32
0:00:32
 0:00:39
0:00:39
 0:53:51
0:53:51
 0:55:58
0:55:58
 0:07:21
0:07:21
 0:00:56
0:00:56
 3:16:41
3:16:41
 0:00:28
0:00:28
 0:00:17
0:00:17
 0:00:34
0:00:34
 1:18:14
1:18:14
 0:03:01
0:03:01
 0:02:04
0:02:04
 0:58:28
0:58:28
 0:01:22
0:01:22
 0:23:40
0:23:40
 0:16:13
0:16:13
 0:12:56
0:12:56
 0:00:40
0:00:40
 0:04:12
0:04:12
 0:05:28
0:05:28