filmov
tv
Constructing a Spatially Resolved Single-cell Atlas of the Mouse Retina with the MERSCOPE Platform

Показать описание
Presented by: Dr. Rui Chen, Ph.D.
Director, ATC Single Cell Genomics Core, Baylor College of Medicine; Professor, HGSC, Departments of Molecular and Human Genetics, Baylor College of Medicine
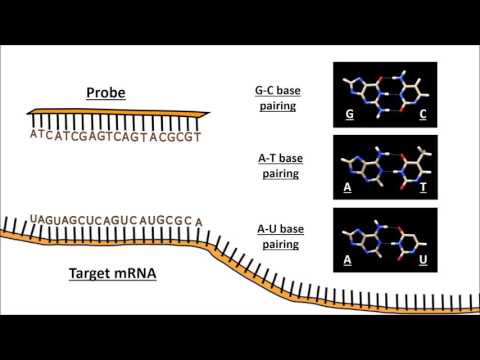
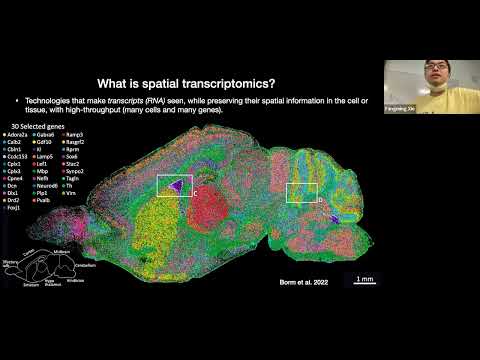
The retina is the light-sensing part of the visual system and is composed of six neuron types and several non-neuronal cell types, with an estimated over 100 subtypes exist based on cell morphology, function, and molecular markers. Recent single-cell transcriptomic studies show that most cell subtypes in the mouse retina can be identified based on their transcriptome. Furthermore, cell subtypes defined by their transcriptome correlated well to results obtained from morphology and function. To gain a thorough understanding of the mechanism of retinal function, the construction of a spatially resolved single-cell atlas of the retina is essential. We performed single-cell spatial transcriptomic profiling on the mouse retina with MERFISH using Vizgen’s MERSCOPE Platform to generate the spatial map of the mouse retina
Webinar Learning Objectives
-Identify why constructing a spatially resolved single-cell atlas of the retina is essential to gain a comprehensive understanding of retinal function
-Discover how a novel procedure was developed to perform single-cell spatial transcriptomic profiling on the mouse retina with MERFISH on Vizgen’s MERSCOPE Platform
-Explore how MERFISH profiles and identifies cell types to enable the spatial mapping of the mouse retina
Director, ATC Single Cell Genomics Core, Baylor College of Medicine; Professor, HGSC, Departments of Molecular and Human Genetics, Baylor College of Medicine
The retina is the light-sensing part of the visual system and is composed of six neuron types and several non-neuronal cell types, with an estimated over 100 subtypes exist based on cell morphology, function, and molecular markers. Recent single-cell transcriptomic studies show that most cell subtypes in the mouse retina can be identified based on their transcriptome. Furthermore, cell subtypes defined by their transcriptome correlated well to results obtained from morphology and function. To gain a thorough understanding of the mechanism of retinal function, the construction of a spatially resolved single-cell atlas of the retina is essential. We performed single-cell spatial transcriptomic profiling on the mouse retina with MERFISH using Vizgen’s MERSCOPE Platform to generate the spatial map of the mouse retina
Webinar Learning Objectives
-Identify why constructing a spatially resolved single-cell atlas of the retina is essential to gain a comprehensive understanding of retinal function
-Discover how a novel procedure was developed to perform single-cell spatial transcriptomic profiling on the mouse retina with MERFISH on Vizgen’s MERSCOPE Platform
-Explore how MERFISH profiles and identifies cell types to enable the spatial mapping of the mouse retina
 0:49:22
0:49:22
 0:49:22
0:49:22
 0:58:04
0:58:04
 1:04:33
1:04:33
 0:26:21
0:26:21
 0:16:52
0:16:52
 1:00:27
1:00:27
 0:05:09
0:05:09
 1:05:16
1:05:16
 0:05:34
0:05:34
 0:05:29
0:05:29
 0:39:36
0:39:36
 0:01:00
0:01:00
 0:32:26
0:32:26
 0:30:44
0:30:44
 1:04:33
1:04:33
 0:57:23
0:57:23
 0:27:12
0:27:12
 2:28:44
2:28:44
 0:26:35
0:26:35
 0:12:12
0:12:12
 2:14:54
2:14:54
 0:06:21
0:06:21
 0:57:21
0:57:21