filmov
tv
Differential Expression Analysis on Array Data from GEO using Limma R

Показать описание
Want to do a Bioinformatics Project from already available datasets online?
If you're a beginner, one of your best options is to perform a Differential Expression Analysis.
This video is about conducting a Differential Expression Analysis on Array data from GEO (Gene Express Omnibus) using the Limma Package in R.
Here, you can see how to search and select a dataset from GEO based on your research question.
Copy-Paste the R script into your RStudio script and run. Adjust the script where necessary according to your dataset and research question.
Refer to the Basic_Instructions_DEAon ArrayDatafromGEO document on how to customize the given workflow and R script for your analysis.
Refer to the RScript_Explanation_DEAonArrayData document to learn more about the R script and Differential Expression Analysis workflow and statistical models.
and/or
In the example shown in this video, I tried to find a dataset of gene expression profiles from Parkinson's patients. It can be mRNA, miRNA, or circRNA but should be profiled using array (i.e. microarray), not by high throughput sequencing (RNA-seq).
This is because the workflow to perform differential expression analysis on RNA-seq data is different from Array data.
there, you can find relevant R scripts and workflows.
Follow TLR4 BioScripts for more Bioinformatics workflows.
#Bioinformatics #BioinformaticsinR #Bioinformaticsforbeginners #Rprogramming #BiomedicalSciences #LifeScience
If you're a beginner, one of your best options is to perform a Differential Expression Analysis.
This video is about conducting a Differential Expression Analysis on Array data from GEO (Gene Express Omnibus) using the Limma Package in R.
Here, you can see how to search and select a dataset from GEO based on your research question.
Copy-Paste the R script into your RStudio script and run. Adjust the script where necessary according to your dataset and research question.
Refer to the Basic_Instructions_DEAon ArrayDatafromGEO document on how to customize the given workflow and R script for your analysis.
Refer to the RScript_Explanation_DEAonArrayData document to learn more about the R script and Differential Expression Analysis workflow and statistical models.
and/or
In the example shown in this video, I tried to find a dataset of gene expression profiles from Parkinson's patients. It can be mRNA, miRNA, or circRNA but should be profiled using array (i.e. microarray), not by high throughput sequencing (RNA-seq).
This is because the workflow to perform differential expression analysis on RNA-seq data is different from Array data.
there, you can find relevant R scripts and workflows.
Follow TLR4 BioScripts for more Bioinformatics workflows.
#Bioinformatics #BioinformaticsinR #Bioinformaticsforbeginners #Rprogramming #BiomedicalSciences #LifeScience
 0:07:41
0:07:41
 0:01:44
0:01:44
 0:09:25
0:09:25
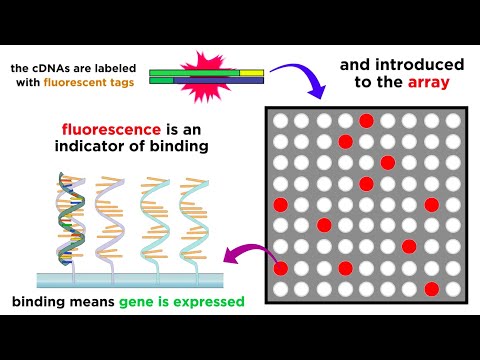 0:08:19
0:08:19
 0:59:25
0:59:25
 0:01:43
0:01:43
 0:17:33
0:17:33
 0:30:36
0:30:36
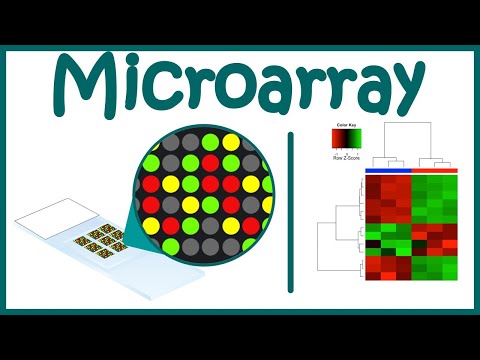 0:09:16
0:09:16
 0:26:33
0:26:33
 0:03:55
0:03:55
 0:16:44
0:16:44
 0:06:14
0:06:14
 0:09:02
0:09:02
 0:52:36
0:52:36
 0:16:33
0:16:33
 0:54:45
0:54:45
 0:28:21
0:28:21
 0:04:16
0:04:16
 0:31:47
0:31:47
 0:57:43
0:57:43
 0:04:12
0:04:12
 1:08:40
1:08:40
 0:02:21
0:02:21