filmov
tv
Molecular cloning overview - techniques & workflow

Показать описание
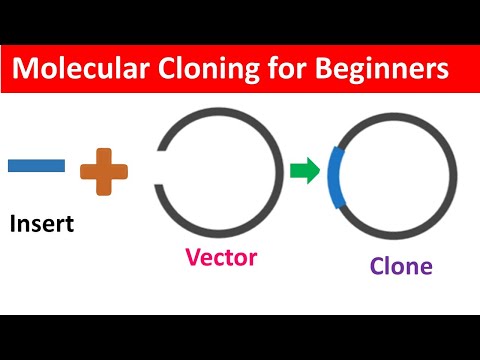
In MOLECULAR CLONING we take a gene* from one place and (most commonly) stick it into a small circular piece of DNA called a PLASMID VECTOR** → stick that into expression cells to use the gene’s instructions to make that protein → get lots of protein you can purify & study (or study how it works in those cells if that’s your goal). I made a video discussing the basics of the process and techniques that it involves from cloning to transformation to that final sequence check. So check it out (if you want)
⠀
⠀
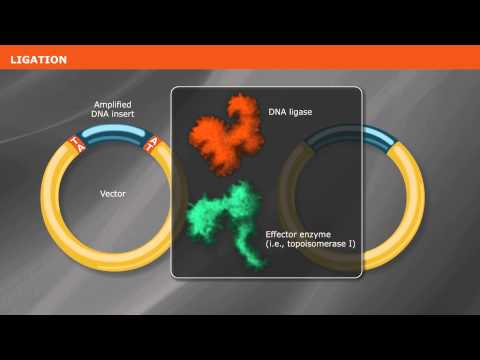
Before you can get that plasmid into the cells you choose, you have to get your gene into that plasmid. The “classic” way to do this is the “cut & paste” with RESTRICTION CLONING. Bacteria have DNA-specific “scissors” called RESTRICTION ENZYMES (aka restriction endonucleases, or REases) that recognize & cut specific “code words” (RESTRICTION SITES aka recognition sequences) written in DNA, which serve as “dotted lines.” With RESTRICTION CLONING, we use these DNA “scissors” to cut an “insert piece” with the gene we want to stick in and a “vector piece” with the vector we want to stick it in with the same pairs of scissors so they have complementary cuts. And then we purify the matching pieces and mix them together, adding a “stitcher” called DNA ligase to seal them up tight. ⠀
⠀
I use a different method, SITE & LIGATION INDEPENDENT CLONING (SLIC). With SLIC cloning we use Polymerase Chain Reaction (PCR) to make lots of copies of (amplify) “insert pieces” & “plasmid pieces.” When we do this copying we add on extra matching bits to the end of the pieces. We then let an enzyme (reaction mediator/speed-upper) chew 1 strand of each of these these ends back a little to leave sticky parts. And then we put them into bacteria to piece them together, fixing the damage. Similar PCR-based methods include Gibson assembly & Golden Gate Assembly. ⠀
⠀
*for proteins, a gene is actually kinda like a “pre-recipe” – proteins are actually made from RNA copies of the DNA recipe. These RNA copies are called messenger RNA (mRNA) and they’re edited (and temporary) copies of the gene – editing involves cutting out regulatory regions (introns) and stitching together the “expressed” regions (exons) in a process called RNA splicing. Our cells do this (and can do it in multiple ways to give you alternative splice products (splice isoforms) – kinda like purposefully skipping a step in a triple-decker cake recipe in order to make a double-decker cake). But bacterial cells don’t – and even if they did, they wouldn’t know which splice isoform to make to know what cake to bake!. So, instead of inserting the gene like it occurs in our DNA, we insert a version of the gene that is complementary to the edited form we want – mature mRNA for the specific isoform of interest. We call this complementary DNA (cDNA). To try to avoid confusion, I’ll use the term “insert” to refer to the cDNA we put it (this is actually a more relevant term anyway because you can clone in *any* DNA - it doesn’t have to be cDNA unless you want to make protein from it.⠀
⠀
**your vector doesn’t have to be a plasmid and sorry in advance for going back-and-forth between “plasmid” & “vector” - a vector is a “vehicle” for transporting your gene into the cells (analogously to how mosquitos can be vectors for malaria, “driving” the malaria DNA into you when they go to suck your blood). Instead of mosquitos, we use things like plasmids, “artificial chromosomes,” and weakened viruses such as adenoviruses (a kind of cold-causers). What kind of vector you use depends largely on the type of cells or organisms you want to get the DNA into and how much DNA you want to stick in.
⠀
I’m going to focus on molecular cloning of plasmids, because that’s probably the most common (and easiest to visualize and stuff). But one of the beautiful biochemical things about molecular cloning is that you can use the same sort of techniques to stick DNA “anywhere.”
There are numerous options including variations & mixtures of:⠀
🔹restriction-enzyme-based: cut & paste⠀
🔹 PCR-based methods: copy just the parts you want and staple together⠀
⠀
regardless of what method you choose you’ll need 2 things:⠀
1️⃣ plasmid vector you want to stick your gene into (destination) ⠀
2️⃣ something containing the gene or “insert” you want to stick into that vector - these days, this insert is usually already inserted into a different plasmid vector just not the one you want so what you need to do is SUBCLONE it → move it from one plasmid to another instead of “traditional” cloning where you’d be moving it from its original location (such as chromosomes inside human cells)
⠀
⠀
⠀
Before you can get that plasmid into the cells you choose, you have to get your gene into that plasmid. The “classic” way to do this is the “cut & paste” with RESTRICTION CLONING. Bacteria have DNA-specific “scissors” called RESTRICTION ENZYMES (aka restriction endonucleases, or REases) that recognize & cut specific “code words” (RESTRICTION SITES aka recognition sequences) written in DNA, which serve as “dotted lines.” With RESTRICTION CLONING, we use these DNA “scissors” to cut an “insert piece” with the gene we want to stick in and a “vector piece” with the vector we want to stick it in with the same pairs of scissors so they have complementary cuts. And then we purify the matching pieces and mix them together, adding a “stitcher” called DNA ligase to seal them up tight. ⠀
⠀
I use a different method, SITE & LIGATION INDEPENDENT CLONING (SLIC). With SLIC cloning we use Polymerase Chain Reaction (PCR) to make lots of copies of (amplify) “insert pieces” & “plasmid pieces.” When we do this copying we add on extra matching bits to the end of the pieces. We then let an enzyme (reaction mediator/speed-upper) chew 1 strand of each of these these ends back a little to leave sticky parts. And then we put them into bacteria to piece them together, fixing the damage. Similar PCR-based methods include Gibson assembly & Golden Gate Assembly. ⠀
⠀
*for proteins, a gene is actually kinda like a “pre-recipe” – proteins are actually made from RNA copies of the DNA recipe. These RNA copies are called messenger RNA (mRNA) and they’re edited (and temporary) copies of the gene – editing involves cutting out regulatory regions (introns) and stitching together the “expressed” regions (exons) in a process called RNA splicing. Our cells do this (and can do it in multiple ways to give you alternative splice products (splice isoforms) – kinda like purposefully skipping a step in a triple-decker cake recipe in order to make a double-decker cake). But bacterial cells don’t – and even if they did, they wouldn’t know which splice isoform to make to know what cake to bake!. So, instead of inserting the gene like it occurs in our DNA, we insert a version of the gene that is complementary to the edited form we want – mature mRNA for the specific isoform of interest. We call this complementary DNA (cDNA). To try to avoid confusion, I’ll use the term “insert” to refer to the cDNA we put it (this is actually a more relevant term anyway because you can clone in *any* DNA - it doesn’t have to be cDNA unless you want to make protein from it.⠀
⠀
**your vector doesn’t have to be a plasmid and sorry in advance for going back-and-forth between “plasmid” & “vector” - a vector is a “vehicle” for transporting your gene into the cells (analogously to how mosquitos can be vectors for malaria, “driving” the malaria DNA into you when they go to suck your blood). Instead of mosquitos, we use things like plasmids, “artificial chromosomes,” and weakened viruses such as adenoviruses (a kind of cold-causers). What kind of vector you use depends largely on the type of cells or organisms you want to get the DNA into and how much DNA you want to stick in.
⠀
I’m going to focus on molecular cloning of plasmids, because that’s probably the most common (and easiest to visualize and stuff). But one of the beautiful biochemical things about molecular cloning is that you can use the same sort of techniques to stick DNA “anywhere.”
There are numerous options including variations & mixtures of:⠀
🔹restriction-enzyme-based: cut & paste⠀
🔹 PCR-based methods: copy just the parts you want and staple together⠀
⠀
regardless of what method you choose you’ll need 2 things:⠀
1️⃣ plasmid vector you want to stick your gene into (destination) ⠀
2️⃣ something containing the gene or “insert” you want to stick into that vector - these days, this insert is usually already inserted into a different plasmid vector just not the one you want so what you need to do is SUBCLONE it → move it from one plasmid to another instead of “traditional” cloning where you’d be moving it from its original location (such as chromosomes inside human cells)
⠀
Комментарии
 0:35:51
0:35:51
 0:06:10
0:06:10
 0:05:49
0:05:49
 0:06:39
0:06:39
 0:08:25
0:08:25
 0:11:07
0:11:07
 0:21:45
0:21:45
 0:02:26
0:02:26
 0:02:46
0:02:46
 0:11:17
0:11:17
 0:16:54
0:16:54
 0:01:42
0:01:42
 0:05:56
0:05:56
 0:16:44
0:16:44
 0:03:07
0:03:07
 0:01:22
0:01:22
 0:08:17
0:08:17
 0:07:33
0:07:33
 0:38:22
0:38:22
 0:14:51
0:14:51
 0:19:18
0:19:18
 0:02:34
0:02:34
 0:10:17
0:10:17
 0:00:50
0:00:50