filmov
tv
How to perform Phylogenetic analysis using MEGA 11 software

Показать описание
#howtoperform #phylogenetic #multiplespecies #mega
In this video, I have how we can build phylogenetic tree of multiple species using MEGA11
🔴If you are a graduate student and trying to learn something new easily, Watch my playlists
If you have queries related to any video/research, please ask in the comments section of the relevant video, so that others can also get benefit from this discussion. [🛑🛑🛑If you are stuck with RNA-seq data analysis or need my help for postdoc/scholarship application: Book a live session: see the below-given link🛑🛑🛑]
🔴 IMPORTANT LINKS 🔴
In this video, I have how we can build phylogenetic tree of multiple species using MEGA11
🔴If you are a graduate student and trying to learn something new easily, Watch my playlists
If you have queries related to any video/research, please ask in the comments section of the relevant video, so that others can also get benefit from this discussion. [🛑🛑🛑If you are stuck with RNA-seq data analysis or need my help for postdoc/scholarship application: Book a live session: see the below-given link🛑🛑🛑]
🔴 IMPORTANT LINKS 🔴
Clint Explains Phylogenetics - There are a million wrong ways to read a phylogenetic tree
Phylogenetic analysis for beginners using MEGA 11 software
1. Phylogenetic analysis of pathogens(lecture - part1) -
Creating a Phylogenetic Tree
Phylogenetic Tree Basics
How to perform Phylogenetic analysis using MEGA 11 software
Cladistics Part 1: Constructing Cladograms
Phylogenetic trees: the basics
Understanding and building phylogenetic trees | High school biology | Khan Academy
How to interpret and understand the results of a phylogenetic tree?
Phylogeny: How We're All Related: Crash Course Biology #17
Interpreting phylogenetic trees
How to perform Phylogenetic Analysis? How to construct phylogenetic tree?
Phylogenetic Tree Construction #iTol #Genomewidestudy
MEGA | How to construct Phylogenetic Tree? | Lecture 14 | Dr. Muhammad Naveed
How do you read Evolutionary Trees?
Notes On Phylogenetic Analysis-Definition &Methods.
Phylogeny and the Tree of Life
How to Construct a Phylogenetic Tree in MEGA 11: A Step-by-Step Guide
Beginners Guide to Clustal Omega | Multiple Sequence Alignment
How To Analyze Phylogenetic Trees | Interpret Bootstrap Values and Sequence Divergence 👨🏻💻🧬...
Steps in Phylogenetic analysis || Phylogenetics lesson #4 || Scoolya
How to do BLAST, Alignment, Phylogenetic tree
How to make phylogenetic tree using mega 11 or Mega XI | Mega software
Комментарии
 0:07:45
0:07:45
 0:11:19
0:11:19
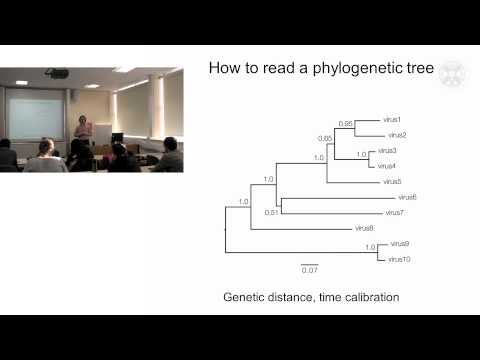 0:07:18
0:07:18
 0:08:51
0:08:51
 0:03:34
0:03:34
 0:13:39
0:13:39
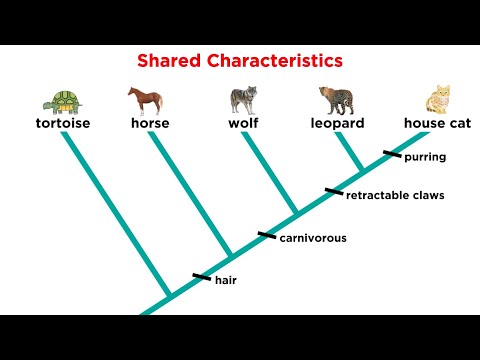 0:10:12
0:10:12
 0:18:20
0:18:20
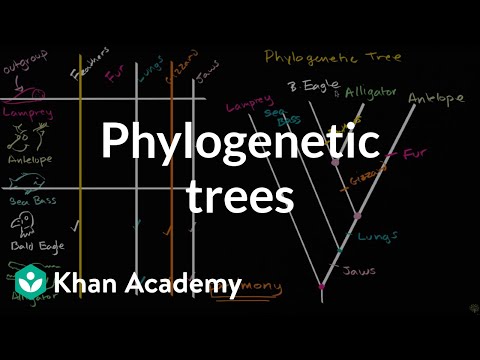 0:10:56
0:10:56
 0:12:23
0:12:23
 0:13:51
0:13:51
 0:22:47
0:22:47
 0:01:41
0:01:41
 0:10:42
0:10:42
 0:10:41
0:10:41
 0:07:36
0:07:36
 0:00:16
0:00:16
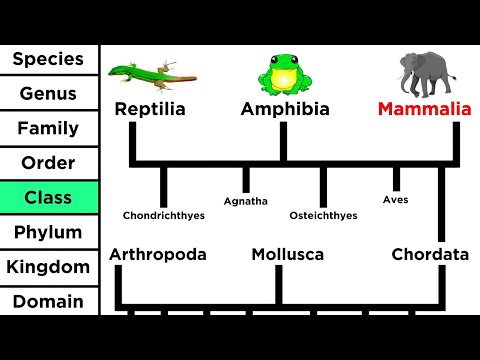 0:11:38
0:11:38
 0:13:48
0:13:48
 0:07:07
0:07:07
 0:18:57
0:18:57
 0:08:19
0:08:19
 0:22:28
0:22:28
 0:13:18
0:13:18