filmov
tv
Standard edgeR Workflow: edgeR Video Tutorial 1

Показать описание
Standard edgeR Workflow: edgeR Video Tutorial 1
Intro to Bioinformatics - EdgeR pt 1 - VDB Computational Biology
Intro to Bioinformatics - EdgeR vs DESeq2 - VDB Computational Biology
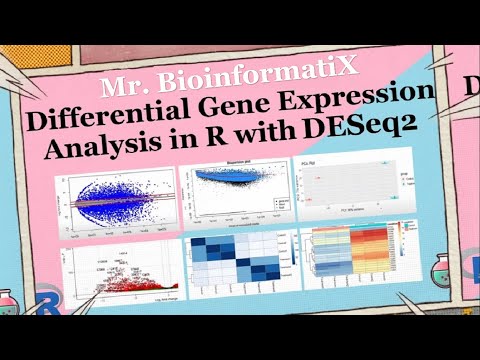
Differential Gene Expression Analysis in R with DESeq2| Bioinformatics Tutorial for Beginners
Multifactor Designs in DESeq2
Differential Expression for RNA-Seq Part 1: Using the limma Bioconductor package
Differential expression analysis
deseq tutorial & visualization. how to plot dispersion estimates
DESeq2 workflow tutorial | Differential Gene Expression Analysis | Bioinformatics 101
DESeq2 Basics Explained | Differential Gene Expression Analysis | Bioinformatics 101
How to install tile edge trim(4 steps)
9.2 Differential expression tests and pathway analysis
STAT115 Chapter 5.2 Differential RNA-seq
The Magic of Multifactor Testing
How Does DESeq2 Work?|Bioinformatics for Beginners|Differential Expression Analysis|Mr BioinformatiX
Tidy Transcriptomics
Stefano Mangiola, Maria Doyle, Workshop 100: A tidy transcriptomics introduction to RNA Seq analyses
Expression and Differential Expression
Differential Expression Analysis with DESeq2
Differential Gene Expression: Bulk RNA Seq with DESEQ2 on the T-BioInfo Platform (Getting Started)
How to use Hyperledger Fabric chaincode events with cc-tools
RNA-seq course: Alignment using TopHat (old)
11 Apr 2019: 'Pachyderm': John Karabaic
B4B: Module 2 - RNAseq analysis workflow
Комментарии
 0:21:59
0:21:59
 0:32:44
0:32:44
 0:17:13
0:17:13
 0:30:36
0:30:36
 0:21:58
0:21:58
 0:12:10
0:12:10
 0:26:33
0:26:33
 0:01:58
0:01:58
 0:19:46
0:19:46
 0:25:32
0:25:32
 0:00:23
0:00:23
 0:14:56
0:14:56
 0:25:20
0:25:20
 0:56:18
0:56:18
 0:27:00
0:27:00
 2:43:03
2:43:03
 0:53:07
0:53:07
 0:57:43
0:57:43
 0:09:56
0:09:56
 0:01:39
0:01:39
 0:09:52
0:09:52
 0:27:39
0:27:39
 0:33:13
0:33:13
 0:13:36
0:13:36