filmov
tv
Tutorial 122 - Segmenting 3D datasets using 3D U-Net

Показать описание
Tutorial 122 - Segmenting 3D datasets using 3D U-Net
3D U-Net for volumetric image segmentation
215 - 3D U-Net for semantic segmentation
V-Net for 3D MRI segmentation
[P070] Semantic Segmentation of 3D Medical Images Through a Kaleidoscope
3D medical image visualization, registration, and segmentation
1. Transform Your Image Segmentation with U-NET
Segmentation and Classification of Brain Tumor from 3D Brain MRI
3D MRI brain segmentation - Made with TensorFlow.js
Tutorial - Intro to segmentation in ITKSNAP
Tutorial 121 - Loading data directly from drive to train U-Net for semantic segmentation
ZeroCostDL4Mic Video #3: Using 3D U-Net for the segmentation of mitochondria from EM data
Modified Open3D which enables detection of 3D dynamic obects using 2D segmentation
Liver Tumor Segmentation and Classification with 3D U-Net in PyTorch | +91-9872993883 for Queries
277 - 3D object segmentation in python
3D Slicer for annotating microscopy data
Segmenting geometries using 3D slicer
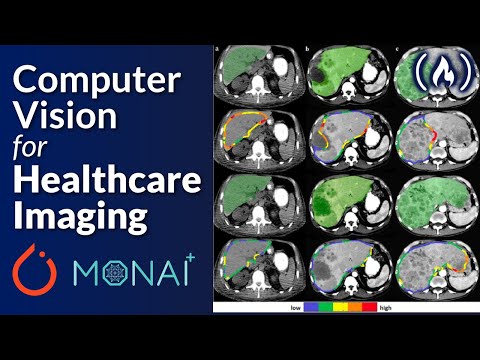
PyTorch and Monai for AI Healthcare Imaging - Python Machine Learning Course
Brain MRI segmentation to mesh file, done in under 20 minutes!
Brain 3D sMRI segmentation into White Matter, Grey Matter, and CSF
[Imagoworks] Deep learning-based fully-automated registration of CBCT and 3D oral scan data
[Tutorial] 3D Cell Analysis
3D Shape Segmentation With Projective Convolutional Networks
aevaSlicer: Grow from Seeds Segmentation
Комментарии
 0:32:18
0:32:18
 0:07:56
0:07:56
 0:50:00
0:50:00
 0:05:29
0:05:29
![[P070] Semantic Segmentation](https://i.ytimg.com/vi/03tWDzQfWlg/hqdefault.jpg) 0:05:08
0:05:08
 0:04:07
0:04:07
 0:04:30
0:04:30
 0:05:02
0:05:02
 0:10:31
0:10:31
 0:07:53
0:07:53
 0:34:33
0:34:33
 0:00:25
0:00:25
 0:00:32
0:00:32
 0:02:39
0:02:39
 0:10:14
0:10:14
 0:32:18
0:32:18
 0:16:31
0:16:31
 5:10:00
5:10:00
 0:04:13
0:04:13
 0:06:19
0:06:19
![[Imagoworks] Deep learning-based](https://i.ytimg.com/vi/XulMpqeFmnQ/hqdefault.jpg) 0:00:26
0:00:26
![[Tutorial] 3D Cell](https://i.ytimg.com/vi/QYbDSz5022s/hqdefault.jpg) 0:04:18
0:04:18
 0:14:02
0:14:02
 0:02:55
0:02:55