filmov
tv
Webinar: New and Improved RNA-Seq Workflows
Показать описание
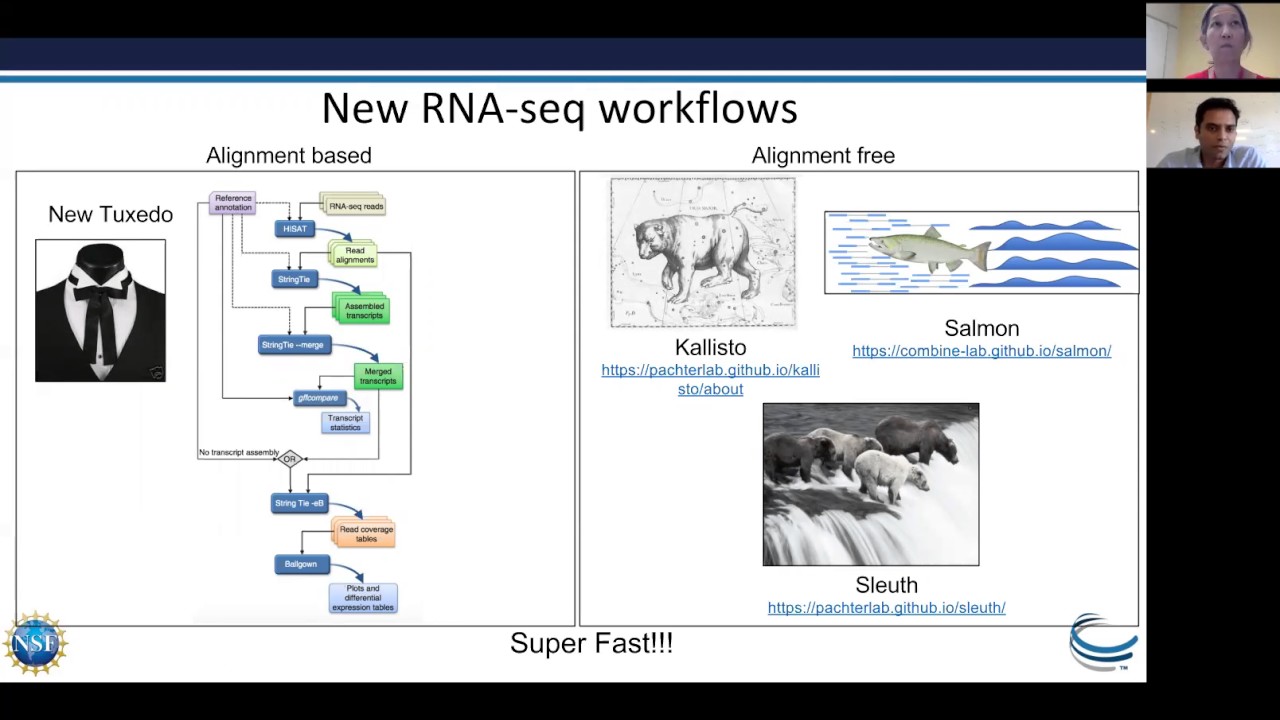
RNA-Seq data analysis has become vital to understanding gene structure and expression patterns in transcriptomic studies. A typical reference genome guided RNA-Seq workflow involves the following steps: quality control of reads, alignment of reads to reference genomes, assembly of reads, quantification, differential expression, and visualization. The current ecosystem of RNA-Seq tools and protocols is vast and diverse. The performance of these tools varies across computing platforms, and hence it becomes critical to choose an analysis protocol that is scalable and accurate when handling large datasets on different computing platforms.
CyVerse provides powerful infrastructure for computational research involving large datasets and complex analyses. Using CyVerse, teams can overcome scaling and complexity challenges to tackle questions that previously were unapproachable.
**Get Started!**
**Make CyVerse work for you!**
Use hundreds of bioinformatics apps and manage data in a simple web interface
**Create a custom cloud-based scientific analysis platform or use a ready-made one for your area of scientific interest.**
**Securely store data for active analyses or sharing with your collaborators.**
**Access discoverable and reusable datasets with metadata features and functions.**
**Exchange, explore, and analyze biological images and their metadata.**
**Follow us!**
Still have questions?
This material is based upon work supported by the National Science Foundation under Grant Nos. DBI-0735191, DBI-1265383, and DBI-1743442.
CyVerse provides powerful infrastructure for computational research involving large datasets and complex analyses. Using CyVerse, teams can overcome scaling and complexity challenges to tackle questions that previously were unapproachable.
**Get Started!**
**Make CyVerse work for you!**
Use hundreds of bioinformatics apps and manage data in a simple web interface
**Create a custom cloud-based scientific analysis platform or use a ready-made one for your area of scientific interest.**
**Securely store data for active analyses or sharing with your collaborators.**
**Access discoverable and reusable datasets with metadata features and functions.**
**Exchange, explore, and analyze biological images and their metadata.**
**Follow us!**
Still have questions?
This material is based upon work supported by the National Science Foundation under Grant Nos. DBI-0735191, DBI-1265383, and DBI-1743442.
Комментарии
 1:03:14
1:03:14
 0:43:36
0:43:36
 0:36:09
0:36:09
 0:50:38
0:50:38
 0:48:19
0:48:19
 0:33:29
0:33:29
 1:03:37
1:03:37
 0:58:58
0:58:58
 1:30:18
1:30:18
 0:30:51
0:30:51
 0:59:32
0:59:32
 1:00:30
1:00:30
 0:59:28
0:59:28
 0:49:57
0:49:57
 1:37:38
1:37:38
 0:42:37
0:42:37
![[WEBINAR] Analysis of](https://i.ytimg.com/vi/y1mFdkDVc-c/hqdefault.jpg) 0:39:06
0:39:06
 1:07:45
1:07:45
 0:33:25
0:33:25
![[Webinar] A Practical](https://i.ytimg.com/vi/LtB6MZx9ra4/hqdefault.jpg) 0:52:46
0:52:46
 0:57:35
0:57:35
 0:07:55
0:07:55
 0:51:46
0:51:46
 1:03:01
1:03:01