filmov
tv
Positive Matrix Factorization for dummies

Показать описание
The theory behind Positive Matrix Factorization (PMF) -a type of factor analysis- explained using soda as an analogy
Positive Matrix Factorization for dummies
Positive Matrix Factorization for dummies (better audio)
How to run and interpret Positive Matrix Factorization (EPA PMF 5.0)
Matrix factorization explained (Part 1)
Matrix Factorization - Numberphile
10701: Non-Negative Matrix Factorization
Non-Negative Matrix Factorization (NMF) | Multiplicative Update Rules By Lee And Seung
Non Negative Matrix Factorization(NMF) - Clustering and Dimensionality Reduction series
How does Netflix recommend movies? Matrix Factorization
Exploring Matrix Factorization with Python: A Step-by-Step Tutorial
PMF Model English version
A Nice Olympiad Exponential Multiplication Problem #short #olympiad #mathematics #maths #exponents
LECTURE 1: Matrix Factorization
On Audio Enhancement via Online Non Negative Matrix Factorization
Eigenvectors and eigenvalues | Chapter 14, Essence of linear algebra
Non-negative Matrix Factorization (NMF) Implementation
Multiply improved positive matrix factorization for source apportionment of || #shortsviral
Lecture 12b: Clustering, NMF, GMMs, and EM
How REAL Men Integrate Functions
Non-negative matrix factorization
Matrix Decompositions
Hey! Be Positive: Nonnegative Matrix Factorization
This trick can make your rubik's cube 2x faster😱🔥#ytshorts#shorts#drcuber
2 Brothers 2 Digit Addition Subtraction | Abacus Classes India | #abacus #youtubeshorts #viralshort
Комментарии
 0:11:00
0:11:00
 0:17:49
0:17:49
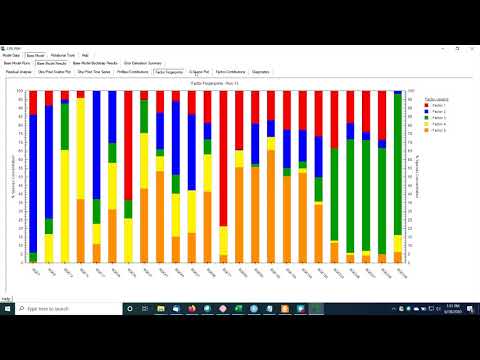 0:22:16
0:22:16
 0:05:01
0:05:01
 0:16:34
0:16:34
 0:10:03
0:10:03
 0:02:45
0:02:45
 0:10:03
0:10:03
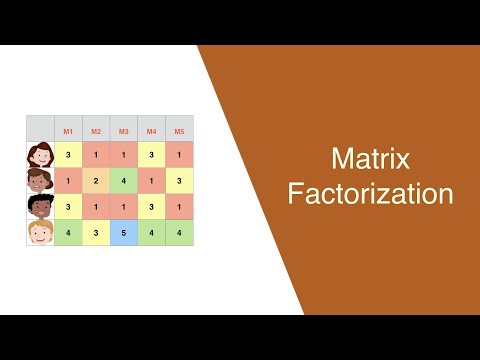 0:32:46
0:32:46
 0:12:55
0:12:55
 3:09:00
3:09:00
 0:00:52
0:00:52
 0:41:38
0:41:38
 0:21:04
0:21:04
 0:17:16
0:17:16
 0:12:23
0:12:23
 0:00:20
0:00:20
 0:50:31
0:50:31
 0:00:35
0:00:35
 0:12:49
0:12:49
 0:22:53
0:22:53
 0:51:21
0:51:21
 0:00:16
0:00:16
 0:00:27
0:00:27