filmov
tv
MS-based proteomics: A short introduction to the core concepts of proteomics and mass spectrometry

Показать описание
A short introduction to the core concepts of MS-based proteomics, which is the use of mass spectrometry to simultaneously measure the abundances, post-translational modifications (PTMs), or interactions of many proteins. I will start by introducing the diverse types of proteomics experiments, how mass spectrometry works, how it is usually combined to do liquid chromatography tandem mass spectrometry (LC-MS-MS), and how it can be used to do either non-targeted or targeted proteomics. I will then move on to the computational aspects of how one can identify what the spectra are and estimate false discovery rate. I explain the differences between label-free and label-based quantification methods and, finally, how statistical analysis is performed to identify statistically significant regulation of proteins, modifications, or interactions.
0:00 Introduction: definition of proteomics, the many flavors, and the steep learning curve
0:41 Experiment types: top-down vs. bottom-up proteomics, quantitative proteomics, phosphoproteomics, PTMs, and affinity purification-mass spectrometry
2:27 Mass spectrometry: a fancy scale, ionization, deflection, detection, mass-to-charge ratio, and peak intensity
3:44 LC-MS-MS: liquid chromatography, tandem mass spectrometry, non-targeted proteomics, and targeted proteomics
5:54 Identification of spectra: de novo peptide sequencing, database search, computed fragment spectra, spectral libraries, peptide spectral matches (PSMs), decoy spectra, false discovery rate, and protein groups
7:54 Quantification: label-free quantification (LFQ), stable isotope labeling, and advantages of comparison within runs vs. between runs
9:33 Statistical analysis: MS-specific analysis software, normalization, and statistical tests
0:00 Introduction: definition of proteomics, the many flavors, and the steep learning curve
0:41 Experiment types: top-down vs. bottom-up proteomics, quantitative proteomics, phosphoproteomics, PTMs, and affinity purification-mass spectrometry
2:27 Mass spectrometry: a fancy scale, ionization, deflection, detection, mass-to-charge ratio, and peak intensity
3:44 LC-MS-MS: liquid chromatography, tandem mass spectrometry, non-targeted proteomics, and targeted proteomics
5:54 Identification of spectra: de novo peptide sequencing, database search, computed fragment spectra, spectral libraries, peptide spectral matches (PSMs), decoy spectra, false discovery rate, and protein groups
7:54 Quantification: label-free quantification (LFQ), stable isotope labeling, and advantages of comparison within runs vs. between runs
9:33 Statistical analysis: MS-specific analysis software, normalization, and statistical tests
Комментарии
 0:10:59
0:10:59
 0:02:33
0:02:33
 0:21:07
0:21:07
 0:06:42
0:06:42
 0:23:07
0:23:07
 0:01:43
0:01:43
 0:03:34
0:03:34
 0:04:42
0:04:42
 0:15:38
0:15:38
 0:02:52
0:02:52
 0:49:54
0:49:54
 0:43:55
0:43:55
 0:24:32
0:24:32
 2:50:52
2:50:52
![[Grow Your Dendrites!]](https://i.ytimg.com/vi/7NEWDmT1sCo/hqdefault.jpg) 0:02:00
0:02:00
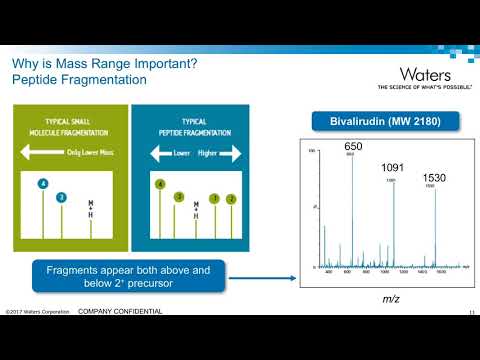 0:19:59
0:19:59
 0:04:51
0:04:51
 0:03:27
0:03:27
 0:32:49
0:32:49
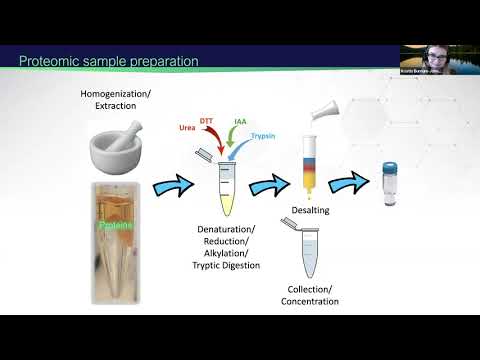 0:43:34
0:43:34
 0:53:28
0:53:28
 0:31:10
0:31:10
 0:50:02
0:50:02
 0:36:27
0:36:27