filmov
tv
Homozygous vs. Heterozygous Samples in qPCR -- Ask TaqMan #23
Показать описание
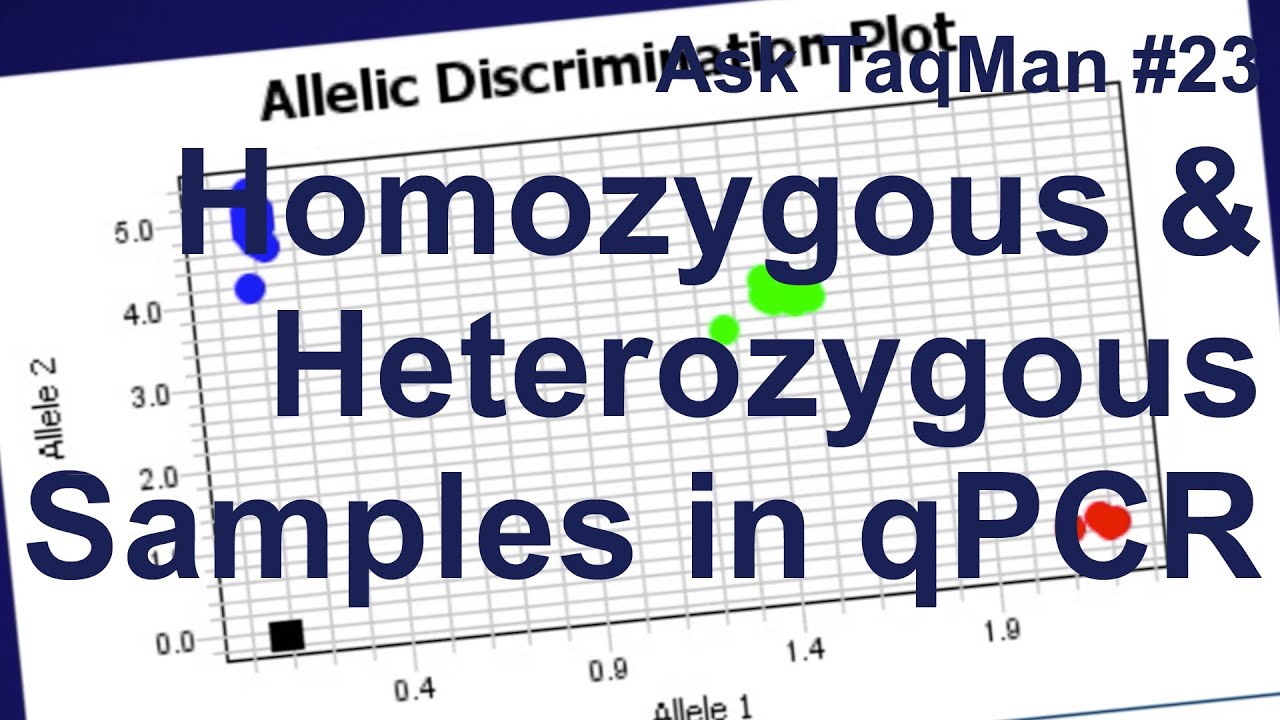
Did you know that TaqMan assays can also be used for SNP Genotyping? While the assays use the same robust TaqMan chemistry, they contain two probes, and thus analysis is different than when using a Gene Expression assay. This prompted our next question, from Melissa Arria from the IVIC in Venezuela, who wondered was seeing amplification from both probes from a sample she knew was homozygous. She asked us: "How can I differentiate real homozygous samples from heterozygous samples?" Great question! But first, let's review how the assay works to understand what we are seeing.
Like a Gene Expression assay, A TaqMan Genotyping assay contains Sequence-specific forward and reverse primers to amplify the polymorphic sequence of interest. However, as we mentioned, the SNP assays are unique since they contain not one, but TWO TaqMan® MGB probes.
-- One probe labeled with VIC® dye detects the Allele 1 sequence
-- One probe labeled with FAM™ dye detects the Allele 2 sequence
Note that all Genotyping assays use the same notation for the context sequence, which is the immediate sequence around the SNP. The first base in brackets is detected by the VIC probe, and the second base by the FAM probe. You can find all this information online for every assay.
The assays are combined with maste rmix and 1-20 ng of purified gDNA, and then amplified. Data can be collected in real-time or off line, as analysis is taken from the end point read of the fluorescence.
The sample will either be homozygous for allele 2, which means we will see mostly VIC dye, homozygous for allele 1, which means we will see mostly FAM signal, or a heterozygous, which means there will be about equal contribution of signal from each dye. When viewed across an entire plate of samples, the data will typically resolve into 3 discrete clusters, assuming enough samples were used to display a normal distribution given the minor allele frequency for your SNP. Note that there are times in which you would not expect to see 3 clusters, such as when the gene is on the X chromosome, if a copy number variation is involved, or if the minor allele frequency is extremely low.
Let's take a closer look at what's happening in each reaction. For example, this sample is a VIC dye homozygous call. However, if we look at the real time data, we'll see some amplification of both probes.
Like a Gene Expression assay, A TaqMan Genotyping assay contains Sequence-specific forward and reverse primers to amplify the polymorphic sequence of interest. However, as we mentioned, the SNP assays are unique since they contain not one, but TWO TaqMan® MGB probes.
-- One probe labeled with VIC® dye detects the Allele 1 sequence
-- One probe labeled with FAM™ dye detects the Allele 2 sequence
Note that all Genotyping assays use the same notation for the context sequence, which is the immediate sequence around the SNP. The first base in brackets is detected by the VIC probe, and the second base by the FAM probe. You can find all this information online for every assay.
The assays are combined with maste rmix and 1-20 ng of purified gDNA, and then amplified. Data can be collected in real-time or off line, as analysis is taken from the end point read of the fluorescence.
The sample will either be homozygous for allele 2, which means we will see mostly VIC dye, homozygous for allele 1, which means we will see mostly FAM signal, or a heterozygous, which means there will be about equal contribution of signal from each dye. When viewed across an entire plate of samples, the data will typically resolve into 3 discrete clusters, assuming enough samples were used to display a normal distribution given the minor allele frequency for your SNP. Note that there are times in which you would not expect to see 3 clusters, such as when the gene is on the X chromosome, if a copy number variation is involved, or if the minor allele frequency is extremely low.
Let's take a closer look at what's happening in each reaction. For example, this sample is a VIC dye homozygous call. However, if we look at the real time data, we'll see some amplification of both probes.
Комментарии
 0:04:37
0:04:37
 0:01:41
0:01:41
 0:05:27
0:05:27
 0:01:47
0:01:47
 0:02:16
0:02:16
 0:02:32
0:02:32
 0:04:16
0:04:16
 0:00:36
0:00:36
 0:02:02
0:02:02
 0:07:06
0:07:06
 0:02:10
0:02:10
 0:09:55
0:09:55
 0:02:07
0:02:07
 0:04:19
0:04:19
 0:05:40
0:05:40
 0:00:09
0:00:09
 0:00:29
0:00:29
 0:00:56
0:00:56
 0:01:45
0:01:45
 0:06:41
0:06:41
 0:00:42
0:00:42
 0:06:31
0:06:31
 0:11:13
0:11:13
 0:29:11
0:29:11