filmov
tv
Basics of Sequence Alignment #Sequence_Alignment #Bioinformatics #DynamicProgramming

Показать описание
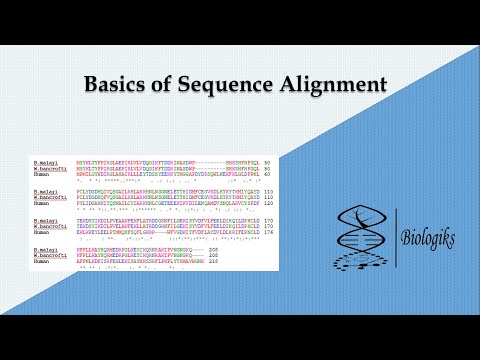
Comparative genomics and genome sequencing allows comparison of organisms at DNA and protein levels, and sequence alignment is a way of arranging the sequences of DNA, RNA, or protein to identify regions of similarity that may be a consequence of functional, structural, or evolutionary relationships between the sequences.
The sequence Comparisons can be used to
- Find evolutionary relationships between organisms
- Identify functionally conserved sequences
- Identify corresponding genes in human and model
- organisms: develop models for human diseases
Therefore, sequence alignment is an important first step toward structural and functional analysis of newly determined sequences to draw functional and evolutionary inference.
In this tutorial we are going to learn about the basics of sequence alignment, and it answers few of the commonly asked questions about sequence alignment. Questions that are addressed in this tutorial includes:
1. Why sequence alignment is performed?
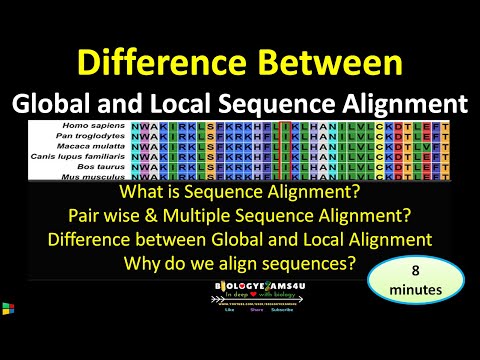
2. What are Global and Local Alignment?
3. How dynamic programming helps in obtaining the optimum alignment, between two sequences?
The sequence Comparisons can be used to
- Find evolutionary relationships between organisms
- Identify functionally conserved sequences
- Identify corresponding genes in human and model
- organisms: develop models for human diseases
Therefore, sequence alignment is an important first step toward structural and functional analysis of newly determined sequences to draw functional and evolutionary inference.
In this tutorial we are going to learn about the basics of sequence alignment, and it answers few of the commonly asked questions about sequence alignment. Questions that are addressed in this tutorial includes:
1. Why sequence alignment is performed?
2. What are Global and Local Alignment?
3. How dynamic programming helps in obtaining the optimum alignment, between two sequences?
Комментарии
 0:16:45
0:16:45
 0:16:26
0:16:26
 0:12:12
0:12:12
 0:11:37
0:11:37
 0:12:38
0:12:38
 0:07:07
0:07:07
 0:08:17
0:08:17
 0:03:39
0:03:39
 0:19:32
0:19:32
 0:09:15
0:09:15
 0:12:04
0:12:04
 0:00:17
0:00:17
 0:00:36
0:00:36
 0:02:37
0:02:37
 0:03:27
0:03:27
 0:00:15
0:00:15
 0:11:33
0:11:33
 0:04:58
0:04:58
 0:05:16
0:05:16
 0:13:05
0:13:05
 0:05:58
0:05:58
 0:00:22
0:00:22
 0:00:39
0:00:39
 0:00:15
0:00:15