filmov
tv
Convert amino acid sequence to 3D protein model in Phyre2

Показать описание
Converting amino acid sequences to 3D protein models in Phyre2.
FREE EBOOK
MORE TO WATCH
SOCIAL MEDIA
WHAT IS DRAWBIOMED
🎨 🧪DrawBioMed is a channel for scientists to learn professional scientific illustrations for their research. Subscribe to @DrawBioMed Scientific Illustrator Jon join this creative scientists community!
GET IN TOUCH
TERMS OF USE
FREE EBOOK
MORE TO WATCH
SOCIAL MEDIA
WHAT IS DRAWBIOMED
🎨 🧪DrawBioMed is a channel for scientists to learn professional scientific illustrations for their research. Subscribe to @DrawBioMed Scientific Illustrator Jon join this creative scientists community!
GET IN TOUCH
TERMS OF USE
Converting Amino Acid Sequence to Peptide Chain 3D Model with Phyre2
Convert amino acid sequence to 3D protein model in Phyre2
Converting Amino Acid Sequences to mRNA
How to Translate mRNA to Amino Acids (DECODING THE GENETIC CODE)
Decode from DNA to mRNA to tRNA to amino acids
How to convert Nucleotide sequence into amino acid sequence?
Decoding the Genetic Code from DNA to mRNA to tRNA to Amino Acid
Expasy Translate Tool | Translate DNA/RNA Sequence to Protein Sequence | @BiologyLectures
Amino Acid Sequence to DNA
Converting RNA sequences to Amino acid sequences
From DNA to protein - 3D
Translating mRNA with a Codon Chart
Nucleotide Sequence Determination from Transcription to Translation
Transcription and Translation - Protein Synthesis From DNA - Biology
Protein Synthesis (Updated)
Bioinformatics; Expasy Translate Tool and Protparam
Bioinformatics; How to retrieve FASTA Protein Sequence
How to translate the CDS into amino acid sequence in GENtle?
How to retrieve an amino acid sequence from nucleotide | How to download protein sequence from NCBI|
Amino acids and protein folding
ExPASy translate tool and Protein Parameters
Working with Translations in SnapGene
How to find nucleotide/protein sequence of a gene in NCBI
Prediction of primary structure of proteins | Expasy translator tool | Bioinformatics
Комментарии
 0:02:24
0:02:24
 0:03:35
0:03:35
 0:03:15
0:03:15
 0:02:56
0:02:56
 0:02:33
0:02:33
 0:02:50
0:02:50
 0:05:28
0:05:28
 0:02:55
0:02:55
 0:02:54
0:02:54
 0:03:10
0:03:10
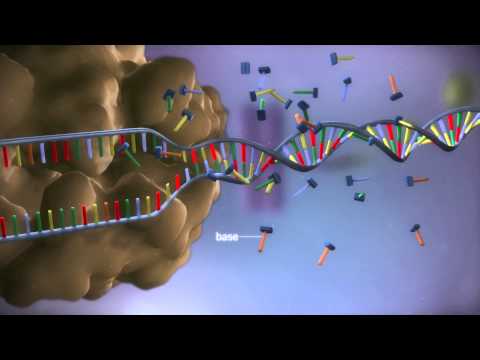 0:02:42
0:02:42
 0:02:47
0:02:47
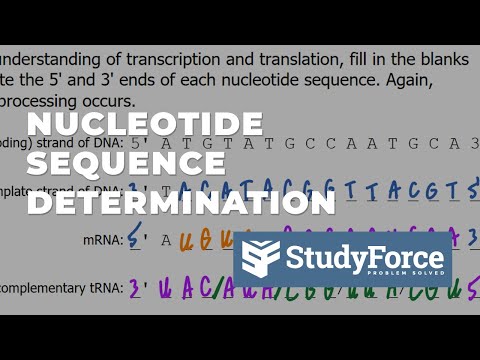 0:05:43
0:05:43
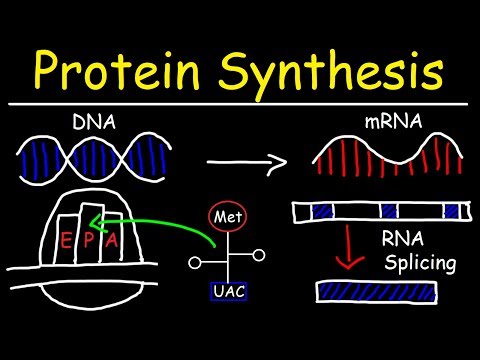 0:10:55
0:10:55
 0:08:47
0:08:47
 0:05:04
0:05:04
 0:02:39
0:02:39
 0:06:10
0:06:10
 0:07:45
0:07:45
 0:11:39
0:11:39
 0:04:29
0:04:29
 0:08:45
0:08:45
 0:12:38
0:12:38
 0:07:13
0:07:13