filmov
tv
How to use NCBI nucleotide BLAST tool

Показать описание
#ncbi #blast #blast #nucleotide
The BLAST search tool can be used to identify matches in gene sequences by comparing the sequence you enter with all recorded sequences in relevant databases. Follow these steps to submit a search and receive results quickly and easily.
Step 1: Open search page
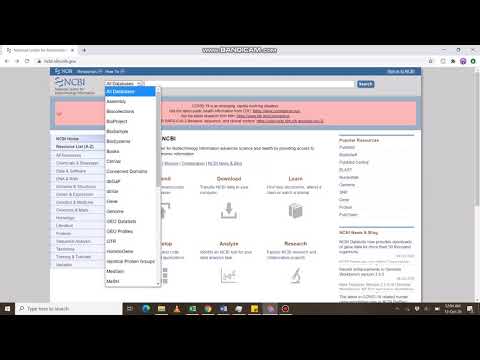
Open the Basic Local Alignment Search Tool page from the National Center for Biotechnology Information website by clicking on BLAST in the Popular Resources menu.
Step 2: Choose program
Choose a BLAST program to run from the Basic BLAST menu; read the short descriptions of each program to figure out which one you should run.
Step 3: Select database
On the program page, select the BLAST database you want to use from the Database pull-down menu under Choose Search Set.
Step 4: Enter sequence
Copy and paste the Accession number, GI number, or FASTA formatted sequence into the large text box under Enter Query Sequence.
Step 5: Submit
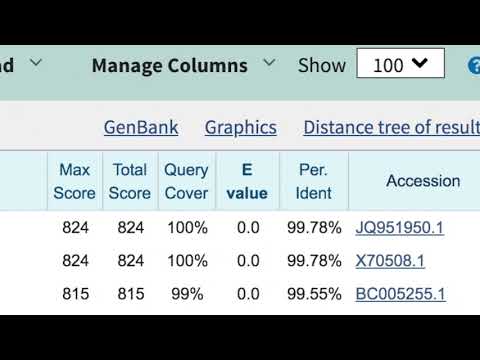
Click on the BLAST button to submit your search; the page will refresh when the results are ready.
blast, how to use blast
how to perform nucleotide blast blastn,
how to use primer blast,
the basic local alignment search tool,
how to run blast on PC,
nucleotide sequence,
nucleotide sequence alignment using blast
NCBI blast,
basic local alignment search tool: nucleotide blast (blastn)
bioinformatics tool
nucleotide database
how to design
nucleotide
use of primer blast
basic local alignment search tool
how to
NCBI nucleotide database
nucleotides
The BLAST search tool can be used to identify matches in gene sequences by comparing the sequence you enter with all recorded sequences in relevant databases. Follow these steps to submit a search and receive results quickly and easily.
Step 1: Open search page
Open the Basic Local Alignment Search Tool page from the National Center for Biotechnology Information website by clicking on BLAST in the Popular Resources menu.
Step 2: Choose program
Choose a BLAST program to run from the Basic BLAST menu; read the short descriptions of each program to figure out which one you should run.
Step 3: Select database
On the program page, select the BLAST database you want to use from the Database pull-down menu under Choose Search Set.
Step 4: Enter sequence
Copy and paste the Accession number, GI number, or FASTA formatted sequence into the large text box under Enter Query Sequence.
Step 5: Submit
Click on the BLAST button to submit your search; the page will refresh when the results are ready.
blast, how to use blast
how to perform nucleotide blast blastn,
how to use primer blast,
the basic local alignment search tool,
how to run blast on PC,
nucleotide sequence,
nucleotide sequence alignment using blast
NCBI blast,
basic local alignment search tool: nucleotide blast (blastn)
bioinformatics tool
nucleotide database
how to design
nucleotide
use of primer blast
basic local alignment search tool
how to
NCBI nucleotide database
nucleotides
 0:11:23
0:11:23
 0:04:47
0:04:47
 0:00:54
0:00:54
 0:32:18
0:32:18
 0:08:24
0:08:24
 0:35:46
0:35:46
 0:00:59
0:00:59
 0:12:38
0:12:38
 0:12:58
0:12:58
 0:57:07
0:57:07
 0:01:29
0:01:29
 0:12:38
0:12:38
 0:06:25
0:06:25
 0:01:31
0:01:31
 0:00:37
0:00:37
 0:12:49
0:12:49
 0:06:35
0:06:35
 0:01:59
0:01:59
 0:23:27
0:23:27
 0:08:44
0:08:44
 0:46:18
0:46:18
 0:11:46
0:11:46
 0:01:29
0:01:29
 0:04:08
0:04:08