filmov
tv
Metagenomics and base modifications from long, short, and linked reads

Показать описание
Speakers:
Christopher Mason, PhD Associate Professor Weill Cornell Medicine WorldQuant (WQ) Scholar & Director, WQ Initiative for Quantitative Prediction
Shuiquan Tang, PhD Scientist, Zymo Research Corp.
Speaker Biography: Dr. Christopher Mason completed his dual B.S. in Genetics and Biochemistry (2001) from University of Wisconsin-Madison, his Ph.D. in Genetics (2006) from Yale University, and then completed post-doctoral training in clinical genetics (2009) at Yale Medical School, and a joint post-doctoral Fellow of Genomics, Ethics, and Law at Yale Law School (2009). He is currently an Associate Professor at Weill Cornell Medicine, with appointments at the Tri-Institutional Program on Computational Biology and Medicine, the Meyer Cancer Center, and the Feil Family Brain and Mind Research Institute.
Dr. Shuiquan Tang is currently a research scientist at Zymo Research. He received his Ph.D. in the Department of Chemical Engineering and Applied Chemistry from the University of Toronto. Dr. Tang joined Zymo Research in 2014, and has since been instrumental in the research and development of Zymo's microbiomics and metagenomics programs. His past experiences include work in the fields of environmental microbiology and biological process engineering, with an emphasis on bioinformatics, metagenomics and anaerobic cultivation. Most recently, Dr. Tang has been involved in the development of Zymo Research's microbiomics portfolio. He has multiple publications in different journals, including Frontiers in Microbiology, Applied & Environmental Microbiology and Philosophical Transactions of the Royal Society of London, Series B, Biological Science.
WEBINAR: Metagenomics and base modifications from long, short, and linked-reads
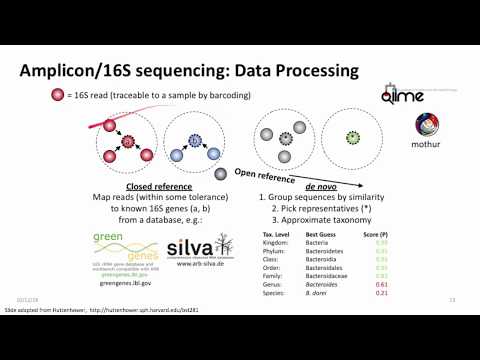
Infectious disease surveillance and monitoring is critical in settings where disease outbreaks and antibiotic resistance can dramatically impact human health. Rapid and accurate detection of human pathogens between closely related strains and across phylogenetic kingdoms is paramount to mitigate disease outbreaks and to diagnose affected individuals. Here, we use demonstrate cross-kingdom species detection by simultaneously sequencing DNA from 8 bacteria and 2 fungi in the ZymoBIOMICS control. We test this on long-read (PacBio, oxford nanopore), short read (Illumina), and linked, long-read (10x Genomics) platforms. We also show that these standard aliquots can be used for benchmarking a variety of tools and also characterize their epigenetic changes (base modifications). These standards are now being used internationally as part of a large-scale effort and improving metagenomics use in the clinic, or “precision metagenomics.”
SPONSORED BY: Zymo Research
Earn PACE Credits:
3. Click Here to get your PACE credits:
LabRoots on Social:
SnapChat: labroots_inc
Christopher Mason, PhD Associate Professor Weill Cornell Medicine WorldQuant (WQ) Scholar & Director, WQ Initiative for Quantitative Prediction
Shuiquan Tang, PhD Scientist, Zymo Research Corp.
Speaker Biography: Dr. Christopher Mason completed his dual B.S. in Genetics and Biochemistry (2001) from University of Wisconsin-Madison, his Ph.D. in Genetics (2006) from Yale University, and then completed post-doctoral training in clinical genetics (2009) at Yale Medical School, and a joint post-doctoral Fellow of Genomics, Ethics, and Law at Yale Law School (2009). He is currently an Associate Professor at Weill Cornell Medicine, with appointments at the Tri-Institutional Program on Computational Biology and Medicine, the Meyer Cancer Center, and the Feil Family Brain and Mind Research Institute.
Dr. Shuiquan Tang is currently a research scientist at Zymo Research. He received his Ph.D. in the Department of Chemical Engineering and Applied Chemistry from the University of Toronto. Dr. Tang joined Zymo Research in 2014, and has since been instrumental in the research and development of Zymo's microbiomics and metagenomics programs. His past experiences include work in the fields of environmental microbiology and biological process engineering, with an emphasis on bioinformatics, metagenomics and anaerobic cultivation. Most recently, Dr. Tang has been involved in the development of Zymo Research's microbiomics portfolio. He has multiple publications in different journals, including Frontiers in Microbiology, Applied & Environmental Microbiology and Philosophical Transactions of the Royal Society of London, Series B, Biological Science.
WEBINAR: Metagenomics and base modifications from long, short, and linked-reads
Infectious disease surveillance and monitoring is critical in settings where disease outbreaks and antibiotic resistance can dramatically impact human health. Rapid and accurate detection of human pathogens between closely related strains and across phylogenetic kingdoms is paramount to mitigate disease outbreaks and to diagnose affected individuals. Here, we use demonstrate cross-kingdom species detection by simultaneously sequencing DNA from 8 bacteria and 2 fungi in the ZymoBIOMICS control. We test this on long-read (PacBio, oxford nanopore), short read (Illumina), and linked, long-read (10x Genomics) platforms. We also show that these standard aliquots can be used for benchmarking a variety of tools and also characterize their epigenetic changes (base modifications). These standards are now being used internationally as part of a large-scale effort and improving metagenomics use in the clinic, or “precision metagenomics.”
SPONSORED BY: Zymo Research
Earn PACE Credits:
3. Click Here to get your PACE credits:
LabRoots on Social:
SnapChat: labroots_inc
 0:59:06
0:59:06
 0:59:06
0:59:06
 0:16:18
0:16:18
 0:12:06
0:12:06
 0:47:26
0:47:26
 0:07:37
0:07:37
 0:22:52
0:22:52
 0:37:44
0:37:44
 1:03:20
1:03:20
 0:30:09
0:30:09
 0:27:53
0:27:53
 0:54:33
0:54:33
 0:41:03
0:41:03
 0:41:47
0:41:47
 0:52:32
0:52:32
 1:39:39
1:39:39
 0:51:54
0:51:54
 0:48:46
0:48:46
 0:11:22
0:11:22
 0:50:27
0:50:27
 1:56:06
1:56:06
 1:03:39
1:03:39
 0:55:30
0:55:30
 0:33:51
0:33:51