filmov
tv
Analyzing Public Datasets 2: Finding the Data

Показать описание
In this new series, we'll learn how to access and analyze public datasets resulting from next-generation sequencing techniques such as Illumina and 454.
This video shows how to find a sample dataset, upload it to Galaxy, and process it for alignment.
This video shows how to find a sample dataset, upload it to Galaxy, and process it for alignment.
Analyzing Public Datasets 2: Finding the Data
Best Places to Find Datasets for Your Projects
Analyzing Public Datasets 6: From start to finish... a TopHat alignment
Analyzing Public Datasets 4: Viewing the Alignment
Master Data Analyst in 2024 with This Proven Roadmap
5 Essential SQL Concepts to Ace Your Data Analysis Interview
Analyzing Public Datasets 3: Performing an Alignment
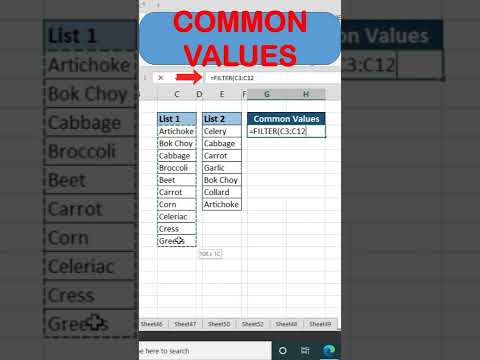
Find Common Values From Two List In Excel @BrainUpp
A Beginners Guide To The Data Analysis Process
Analyzing Public Datasets 1: Introduction to Galaxy
Analyzing Public Datasets 7: An Introduction to IGV (genomic)
2 websites for finding ML Datasets! 📈🤖 #programming #coding #machinelearning
Analyzing Public Datasets 5: Introduction to IGV (cDNA)
Life as a Data Analyst #shorts
10 Free Dataset Resources for Your Next Project!
Loading, Viewing, working with an R dataset (basics)
Don’t Become a Data Scientist If
My 3 go-to places to find interesting real-life datasets
How to compare two lists to find missing values in excel - Excel Tips and Tricks
Real Time Power BI Project, Blinkit Analysis #powerbi #powerbidashboard #dataanalyst
Explore data from chronic liver disease and find other related public datasets.
🔥Salary of a Data Scientist | How Much Do Data Scientists Make? | Intellipaat #Shorts #DataScientist...
Google Cloud Public Datasets Program Overview
5 FREE websites with datasets for your data projects: 1
Комментарии
 0:08:59
0:08:59
 0:07:44
0:07:44
 0:09:25
0:09:25
 0:06:23
0:06:23
 0:00:05
0:00:05
 0:00:05
0:00:05
 0:04:12
0:04:12
 0:00:39
0:00:39
 0:10:20
0:10:20
 0:03:38
0:03:38
 0:05:02
0:05:02
 0:00:28
0:00:28
 0:08:54
0:08:54
 0:00:14
0:00:14
 0:05:47
0:05:47
 0:11:21
0:11:21
 0:00:27
0:00:27
 0:00:54
0:00:54
 0:00:31
0:00:31
 0:00:15
0:00:15
 0:33:07
0:33:07
 0:00:25
0:00:25
 0:01:38
0:01:38
 0:00:14
0:00:14