filmov
tv
For bioinformatics, which language should I learn first?

Показать описание
Whether you are a wet lab biologist wanting to add some bioinformatics skills to your toolbox or if you are trying to get all the way into bioinformatics and make your own software, I have some concrete recommendations about what programming languages to start with. I focus on R, Python, and Bash for beginners, and I also consider in what circumstances you should consider adding other languages, including C, C++, Ruby, Java, PHP, JavaScript, and Matlab.
For bioinformatics, which language should I learn first?
Which Programming Language Should I Learn first as Bioinformatics Beginner || Python Linux & R
Best Programming Languages for Bioinformatics | Must-Have Skills for Computational Biology #biotech
Most Popular Programming Languages For Bioinformatics || By NaveeNBioinformaticS
Top 7 Programming Languages Every Biologist Must Know! #bioinformatics #coding #programming
The First 2 Programming Languages You Should Learn for Bioinformatics
R versus Python for bioinformatics
Learning BIOINFORMATICS in 2023 - What I would do differently! - Genomics with Georgia
Python Vs R (funny!)
Python vs Julia
Bioinformatics for Beginners: Essential Skills for Bioinformatics| Roadmap to learn #Bioinformatics
Don’t Do Bioinformatics/Data Science. Here is why #bioinformatics
Top Programing Language For Bioinformatics
What is Bioinformatics?
Python vs R language best for Bioinformatics
R vs Python for Bioinformatics
R vs Python| R vs. Python for Bioinformatics and Biology| Programming for Biology
Beginners Python for Bioinformatics
5 Steps to Transitioning Into Bioinformatics As A Bio Student
Beginners R for Bioinformatics
Which is Faster for Bioinformatics Python or R
HIGHEST PAID HEALTHCARE WORKERS 💰 (that aren't medical doctors) #shorts
Most popular programming languages for Bioinformatics (1998-2019)
PROGRAMMING for Biologists| Roadmap to Learn Programming for biologist #bioinformatics #biotech
Комментарии
 0:09:43
0:09:43
 0:07:42
0:07:42
 0:00:41
0:00:41
 0:05:31
0:05:31
 0:00:58
0:00:58
 0:11:21
0:11:21
 0:05:08
0:05:08
 0:13:30
0:13:30
 0:03:23
0:03:23
 0:07:10
0:07:10
 0:06:05
0:06:05
 0:00:09
0:00:09
 0:01:13
0:01:13
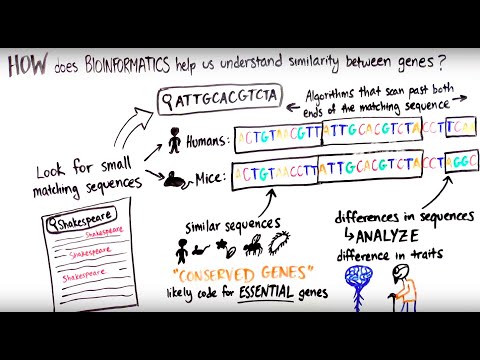 0:05:35
0:05:35
 0:04:51
0:04:51
 0:04:54
0:04:54
 0:03:23
0:03:23
 0:00:58
0:00:58
 0:28:19
0:28:19
 0:00:55
0:00:55
 0:06:30
0:06:30
 0:00:14
0:00:14
 0:01:07
0:01:07
 0:00:43
0:00:43