filmov
tv
Transcription Termination in Eukaryotes

Показать описание
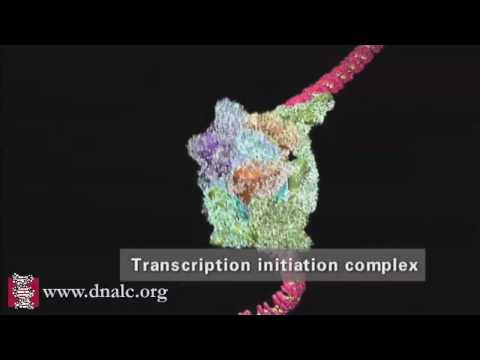
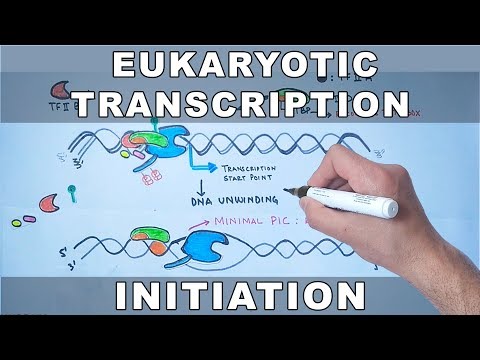
Eukaryotic transcription is the elaborate process that eukaryotic cells use to copy genetic information stored in DNA into units of transportable complementary RNA replica.[1] Gene transcription occurs in both eukaryotic and prokaryotic cells. Unlike prokaryotic RNA polymerase that initiates the transcription of all different types of RNA, RNA polymerase in eukaryotes (including humans) comes in three variations, each translating a different type of gene. A eukaryotic cell has a nucleus that separates the processes of transcription and translation. Eukaryotic transcription occurs within the nucleus where DNA is packaged into nucleosomes and higher order chromatin structures. The complexity of the eukaryotic genome necessitates a great variety and complexity of gene expression control.
The termination of transcription of pre-rRNA genes by polymerase Pol I is performed by a system that needs a specific transcription termination factor.[3] The mechanism used bears some resemblance to the rho-dependent termination in prokaryotes.[36] Eukaryotic cells contain hundreds of ribosomal DNA repeats, sometimes distributed over multiple chromosomes. Termination of transcription occurs in the ribosomal intergenic spacer region that contains several transcription termination sites upstream of a Pol I pausing site. Through a yet unknown mechanism, the 3’-end of the transcript is cleaved, generating a large primary rRNA molecule that is further processed into the mature 18S, 5.8S and 28S rRNAs.
As Pol II reaches the end of a gene, two protein complexes carried by the CTD, CPSF (cleavage and polyadenylation specificity factor) and CSTF (cleavage stimulation factor), recognize the poly-A signal in the transcribed RNA.[35] Poly-A-bound CPSF and CSTF recruit other proteins to carry out RNA cleavage and then polyadenylation. Poly-A polymerase adds approximately 200 adenines to the cleaved 3’ end of the RNA without a template.[35] The long poly-A tail is unique to transcripts made by Pol II.
In the process of terminating transcription by Pol I and Pol II, the elongation complex does not dissolve immediately after the RNA is cleaved. The polymerase continues to move along the template, generating a second RNA molecule associated with the elongation complex.[1] Two models have been proposed to explain how termination is achieved at last.[35] The allosteric model states that when transcription proceeds through the termination sequence, it causes disassembly of elongation factors and/or an assembly of termination factors that cause conformational changes of the elongation complex.[36][37] The torpedo model suggests that a 5' to 3' exonuclease degrades the second RNA as it emerges from the elongation complex. Polymerase is released as the highly processive exonuclease overtakes it. It is proposed that an emerging view will express a merge of these two models.
The termination of transcription of pre-rRNA genes by polymerase Pol I is performed by a system that needs a specific transcription termination factor.[3] The mechanism used bears some resemblance to the rho-dependent termination in prokaryotes.[36] Eukaryotic cells contain hundreds of ribosomal DNA repeats, sometimes distributed over multiple chromosomes. Termination of transcription occurs in the ribosomal intergenic spacer region that contains several transcription termination sites upstream of a Pol I pausing site. Through a yet unknown mechanism, the 3’-end of the transcript is cleaved, generating a large primary rRNA molecule that is further processed into the mature 18S, 5.8S and 28S rRNAs.
As Pol II reaches the end of a gene, two protein complexes carried by the CTD, CPSF (cleavage and polyadenylation specificity factor) and CSTF (cleavage stimulation factor), recognize the poly-A signal in the transcribed RNA.[35] Poly-A-bound CPSF and CSTF recruit other proteins to carry out RNA cleavage and then polyadenylation. Poly-A polymerase adds approximately 200 adenines to the cleaved 3’ end of the RNA without a template.[35] The long poly-A tail is unique to transcripts made by Pol II.
In the process of terminating transcription by Pol I and Pol II, the elongation complex does not dissolve immediately after the RNA is cleaved. The polymerase continues to move along the template, generating a second RNA molecule associated with the elongation complex.[1] Two models have been proposed to explain how termination is achieved at last.[35] The allosteric model states that when transcription proceeds through the termination sequence, it causes disassembly of elongation factors and/or an assembly of termination factors that cause conformational changes of the elongation complex.[36][37] The torpedo model suggests that a 5' to 3' exonuclease degrades the second RNA as it emerges from the elongation complex. Polymerase is released as the highly processive exonuclease overtakes it. It is proposed that an emerging view will express a merge of these two models.
Комментарии
 0:05:14
0:05:14
 0:05:32
0:05:32
 0:16:35
0:16:35
 0:08:45
0:08:45
 0:15:38
0:15:38
 1:25:29
1:25:29
 0:10:24
0:10:24
 0:01:54
0:01:54
 0:02:30
0:02:30
 0:15:24
0:15:24
 0:05:21
0:05:21
 0:04:01
0:04:01
 0:04:16
0:04:16
 0:10:55
0:10:55
 0:07:32
0:07:32
 0:06:27
0:06:27
 0:09:01
0:09:01
 0:03:18
0:03:18
 0:23:48
0:23:48
 0:05:26
0:05:26
 0:02:08
0:02:08
 0:16:13
0:16:13
 0:10:23
0:10:23
 0:05:01
0:05:01