filmov
tv
ImageJ plugin: Semi-automatic cell counting (with colocalization / categorization). Object based.
Показать описание
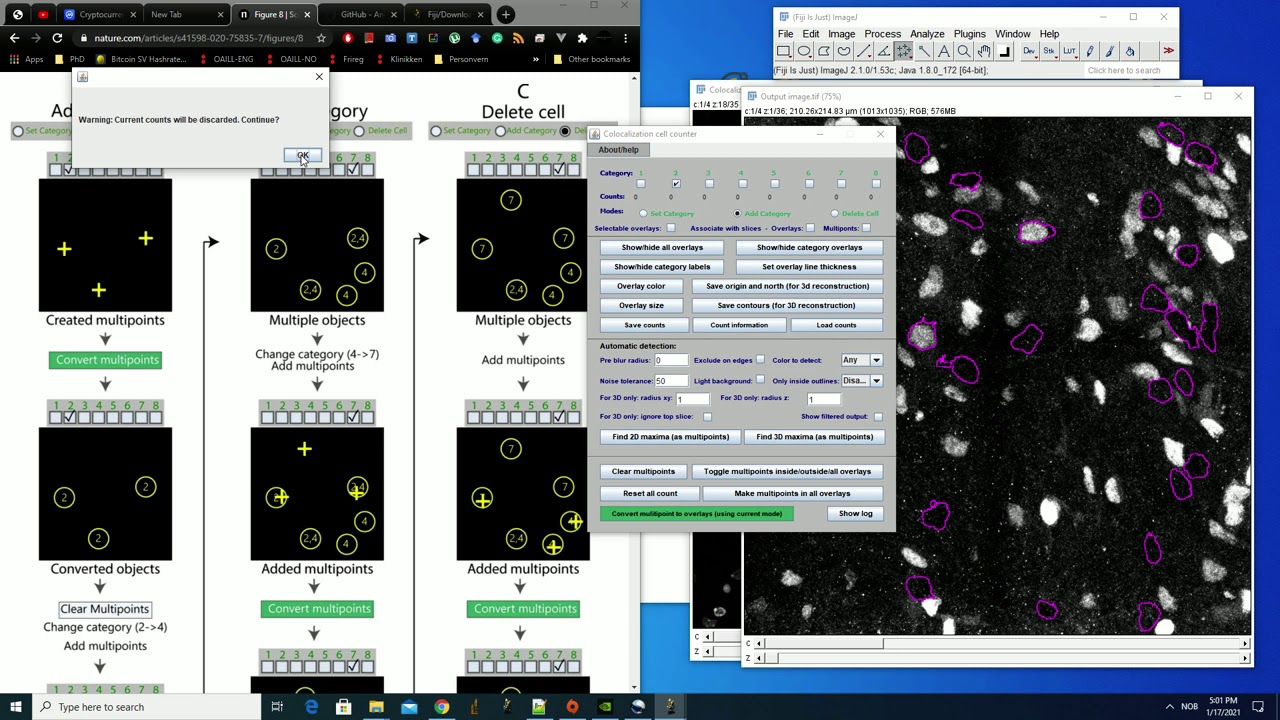
new ImageJ plugin: A versatile toolbox for semi-automatic cell-by-cell object-based colocalization analysis
Features:
ImageJ plugin 1: Colocalization Image Creator:
*Pre-process multichannel Z-stack (or 2D) microscopy images into a visual format for faster, simpler, and more accurate colocalization analysis.
*Designed to help avoid common colocalization analysis artifacts and errors.
*Can transform Z-stack 3D data into a specialized 2D Z-projection where Z-projection colocalization artifacts are removed/reduced. This simplifies the analysis of 3D colocalization data.
ImageJ plugin 2: Colocalization Object Counter:
*Quantity (count) cells/objects in a semi-automatic manner.
*Assign, classify and keep track of multichannel signal presence/absence (colocalization analysis) for each cell/object.
*Tools for subsequent 3D modeling/representation of data: draw tissue contours and indicate image-series global XY-origin.
*Save data, load data, and export data to Excel.
Custom Excel macro-file:
*Import data from Colocalization Object Counter
*Analyze and edit data from image series.
*Export combined image series data to Matlab for 3D modeling
Custom Matlab script:
*3D visualize cells according to colocalization data
*3D visualize tissue contours
I hope the community will appreciate our work. The ImageJ plugin 1 might be somewhat hard to understand how to use effectively (though we hope not), but ImageJ plugin 2 should be very simple and useful to the broader community. I found a good cell counting tool for ImageJ lacking, so maybe this plugin (and the other) can be included as a standard part of FIJI.
Sincerely,
Anders Lunde, PhD
University of Oslo, Norway
Features:
ImageJ plugin 1: Colocalization Image Creator:
*Pre-process multichannel Z-stack (or 2D) microscopy images into a visual format for faster, simpler, and more accurate colocalization analysis.
*Designed to help avoid common colocalization analysis artifacts and errors.
*Can transform Z-stack 3D data into a specialized 2D Z-projection where Z-projection colocalization artifacts are removed/reduced. This simplifies the analysis of 3D colocalization data.
ImageJ plugin 2: Colocalization Object Counter:
*Quantity (count) cells/objects in a semi-automatic manner.
*Assign, classify and keep track of multichannel signal presence/absence (colocalization analysis) for each cell/object.
*Tools for subsequent 3D modeling/representation of data: draw tissue contours and indicate image-series global XY-origin.
*Save data, load data, and export data to Excel.
Custom Excel macro-file:
*Import data from Colocalization Object Counter
*Analyze and edit data from image series.
*Export combined image series data to Matlab for 3D modeling
Custom Matlab script:
*3D visualize cells according to colocalization data
*3D visualize tissue contours
I hope the community will appreciate our work. The ImageJ plugin 1 might be somewhat hard to understand how to use effectively (though we hope not), but ImageJ plugin 2 should be very simple and useful to the broader community. I found a good cell counting tool for ImageJ lacking, so maybe this plugin (and the other) can be included as a standard part of FIJI.
Sincerely,
Anders Lunde, PhD
University of Oslo, Norway
Комментарии
 0:35:52
0:35:52
 0:06:56
0:06:56
 0:01:07
0:01:07
 0:04:48
0:04:48
 1:03:36
1:03:36
 0:06:11
0:06:11
 0:07:47
0:07:47
 0:21:06
0:21:06
 0:02:49
0:02:49
 0:10:43
0:10:43
 0:00:55
0:00:55
 0:04:45
0:04:45
 0:01:03
0:01:03
 0:10:34
0:10:34
 0:12:21
0:12:21
 0:02:48
0:02:48
 0:04:47
0:04:47
 0:03:41
0:03:41
 0:02:54
0:02:54
 0:07:20
0:07:20
 0:07:03
0:07:03
 0:08:27
0:08:27
 0:05:04
0:05:04
 0:02:54
0:02:54