filmov
tv
ncbi-blast+ tutorial, blast from the command line (part 1)

Показать описание
This video demonstrates how to use ncbi-blast+. the following commands
are used in the video and the next video:
makeblastdb: used to create a blastdb from a fasta file
blastn: used to do BLAST alignment on a query sequence of nucleic acid against a database of nucleic acid
grep [pattern to search for] [file]
tail -N file (where N is the number of lines from the bottom you want to return)
less [file] display contents of file to terminal
are used in the video and the next video:
makeblastdb: used to create a blastdb from a fasta file
blastn: used to do BLAST alignment on a query sequence of nucleic acid against a database of nucleic acid
grep [pattern to search for] [file]
tail -N file (where N is the number of lines from the bottom you want to return)
less [file] display contents of file to terminal
How to Use BLAST for Finding and Aligning DNA or Protein Sequences
How to run NCBI blast | NCBI blast tutorial | How to perform or use blast practically
NCBI Blast Tutorial
Analyzing Gene Sequence Results with BLAST
NCBI Blast Tutorial - Analyzing gene sequence results
BLAST Tutorial
ncbi-blast+ tutorial, blast from the command line (part 1)
NCBI Ig Blast tutorial I xgene and proteinx
How to perform local blast on computer? Complete tutorial installation to run.
How to Use NCBI Blast
How to use NCBI BLAST| What is BLAST| NCBI Tutorial|Protein and DNA sequence alignment tool
How to Use the NCBI’s Bioinformatics Tools and Databases
NCBI BLAST | Annotation Tutorial Video 2
NCBI BLAST Tutorial - Part 1
Introduction to NCBI: Tutorial 1- General use and basic BLAST
What is BLAST? - Tutorial & Interpretation
How to perform blast analysis on NCBI | Tutorial 5
(Bioinformatics) NCBI PSI-BLAST Tutorial | Power BLAST Type | Bioinformatic Tutorials
Use of NCBI BLAST for two sequences (pairwise alignment)
how to do blast in NCBI||bioinformatics practical blast||in easy way#from undefined learning
BLAST in Bioinformatics: Nucleotide & Protein Alignment Made Easy! | NCBI BLAST Tutorial
How to use NCBI BLAST || NCBI BLAST Tutorial || NCBI BLAST Practical
DNA Master Tutorial 5 - How To Blast All Genes to NCBI Database!
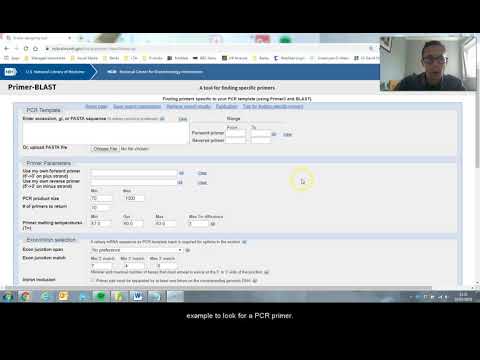
How to use Primer-Blast to create PCR primers
Комментарии
 0:12:38
0:12:38
 0:11:46
0:11:46
 0:07:55
0:07:55
 0:08:24
0:08:24
 0:07:28
0:07:28
 0:01:59
0:01:59
 0:16:42
0:16:42
 0:06:17
0:06:17
 0:05:11
0:05:11
 0:01:55
0:01:55
 0:23:44
0:23:44
 0:11:23
0:11:23
 0:15:23
0:15:23
 0:01:12
0:01:12
 0:16:12
0:16:12
 0:11:41
0:11:41
 0:05:14
0:05:14
 0:06:17
0:06:17
 0:00:39
0:00:39
 0:06:01
0:06:01
 0:13:02
0:13:02
 0:11:44
0:11:44
 0:05:08
0:05:08
 0:05:12
0:05:12