filmov
tv
Downloading Macromolecular Sequences from the NCBI Database
Показать описание
An important step in providing sequence database access was the development of
Web pages that allow queries to be made of the major sequence databases (GenBank, EMBL, etc.). An early example of this technology at NCBI was a menu driven program called GEN-INFO developed by D. Benson, D. Lipman, and colleagues.
This program searched rapidly through previously indexed sequence databases for entries that matched a biologist’s query.
Subsequently, a derivative program called ENTREZ with a simple window-based interface, and eventually a Web-based interface, was developed at NCBI. The idea behind these programs was to provide an easy-to-use interface with a flexible search
procedure to the sequence databases.
Sequence entries in the major databases have additional information about the sequence included with the sequence entry, such as accession or index number, name and alternative names for the sequence, names of relevant genes, types of
regulatory sequences, the source organism, references, and known mutations.
Procedure:
Step - 1: At First, We need to go to the NCBI website
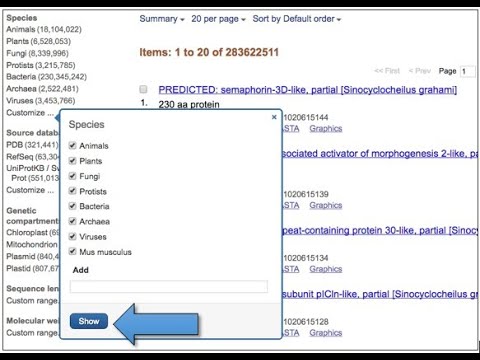
Step – 2: Then, we can see a search bar on the top of the website along with a drop down box from which we can select the type of the sequence databases (such as Protein, Nucleotide, Genome etc).
Step – 3: After that, we can select Protein from the dropdown box and type any example of Protein (such as Cellulose, Elastin etc) on the search bar to search a Protein Sequence.
Or, we can select Nucleotide from the dropdown box and type example of any organism we want to study (such as Elephant, Tigers, Cat etc) on the search bar to search Nucleotide sequence.
Step – 4: After clicking on the search button we can get large number of data in the form of search result according to the given query.
Step – 5: Therefore, we can filter out the search results according to our requirements so that we can get the required data.
Step - 6: After opening the next window we can click on the FASTA button to get the sequence in the FASTA file format.
Step – 7: Finally, we can copy the whole sequence in the Notepad and type ctrl+s.
Web pages that allow queries to be made of the major sequence databases (GenBank, EMBL, etc.). An early example of this technology at NCBI was a menu driven program called GEN-INFO developed by D. Benson, D. Lipman, and colleagues.
This program searched rapidly through previously indexed sequence databases for entries that matched a biologist’s query.
Subsequently, a derivative program called ENTREZ with a simple window-based interface, and eventually a Web-based interface, was developed at NCBI. The idea behind these programs was to provide an easy-to-use interface with a flexible search
procedure to the sequence databases.
Sequence entries in the major databases have additional information about the sequence included with the sequence entry, such as accession or index number, name and alternative names for the sequence, names of relevant genes, types of
regulatory sequences, the source organism, references, and known mutations.
Procedure:
Step - 1: At First, We need to go to the NCBI website
Step – 2: Then, we can see a search bar on the top of the website along with a drop down box from which we can select the type of the sequence databases (such as Protein, Nucleotide, Genome etc).
Step – 3: After that, we can select Protein from the dropdown box and type any example of Protein (such as Cellulose, Elastin etc) on the search bar to search a Protein Sequence.
Or, we can select Nucleotide from the dropdown box and type example of any organism we want to study (such as Elephant, Tigers, Cat etc) on the search bar to search Nucleotide sequence.
Step – 4: After clicking on the search button we can get large number of data in the form of search result according to the given query.
Step – 5: Therefore, we can filter out the search results according to our requirements so that we can get the required data.
Step - 6: After opening the next window we can click on the FASTA button to get the sequence in the FASTA file format.
Step – 7: Finally, we can copy the whole sequence in the Notepad and type ctrl+s.