filmov
tv
Loops At Command Line

Показать описание
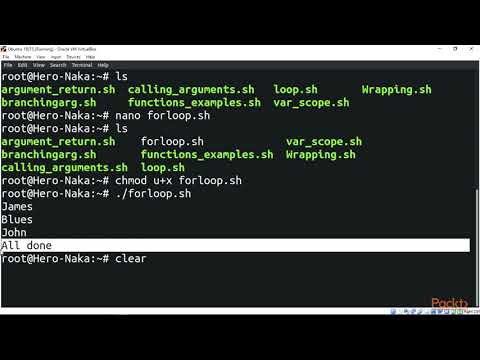
Loops can provide an efficient way to perform iterative actions at command line. This video provides a brief introduction to writing and executing "for loops" at command line, with a specific example of using Bowtie2 software to perform DNA sequence alignments for a series of files.
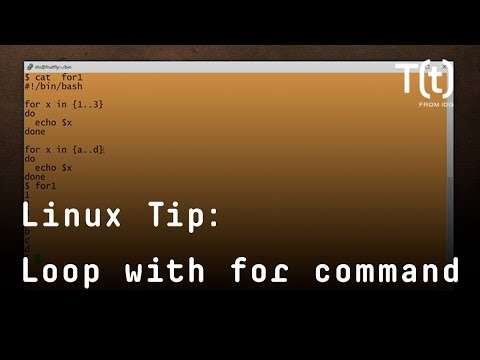
Simple for loops on the Linux command line
Command Line: For Loops
Command Line: While Loops
Linux Shell Scripting: Automating Command Line Tasks: Loops – for and while Loops | packtpub.com
2-Minute Linux Tips: How to run command loops
Shell Scripting - For Loops
Linux Command Line Basics - A directory loop !
How to end a loop on your command line (packet tracer)
COMP1511 Week 4 Lecture 2
Shell scripting: While loop and read command: Read file line by line
WIN7: Windows for Linux Admins: Command Line Users/groups and For loops
Intro to Programming: Loops
Shell Scripting Tutorial for Beginners 20 - use FOR loop to execute commands
PHP Tutorial: Command Line Scripts [part 03]: Infinite Loops
Learning Awk Is Essential For Linux Users
'Babashka: a meta-circular Clojure interpreter for the command line' by Michiel Borkent
Shell Scripting Tutorial for Beginners 18 - UNTIL loop
8 Hours Terminal Command Line
linux administrator:53 for loop using command line arguments
How To Make Command Prompt Open In A Loop FOREVER
The HARDEST part about programming 🤦♂️ #code #programming #technology #tech #software #developer...
Never say 'If' writing a Bash script! (Exit codes & logical operators)
#tree #command #prompt #command #loop. @CodeWithNaman
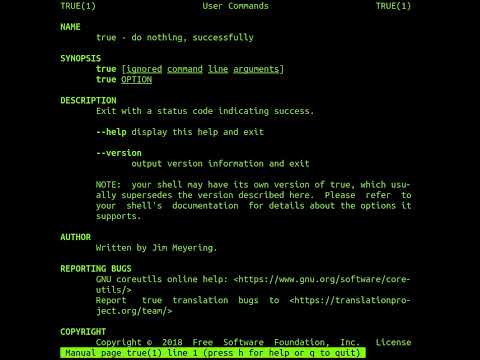
The 'true' Command In Linux
Комментарии
 0:04:59
0:04:59
 0:02:40
0:02:40
 0:02:25
0:02:25
 0:05:02
0:05:02
 0:04:18
0:04:18
 0:09:22
0:09:22
 0:04:28
0:04:28
 0:00:53
0:00:53
 1:53:43
1:53:43
 0:02:54
0:02:54
 0:08:14
0:08:14
 0:01:41
0:01:41
 0:06:46
0:06:46
 0:03:19
0:03:19
 0:20:02
0:20:02
 0:39:51
0:39:51
 0:04:22
0:04:22
 8:07:15
8:07:15
 0:09:12
0:09:12
 0:03:26
0:03:26
 0:00:28
0:00:28
 0:14:50
0:14:50
 0:00:15
0:00:15
 0:00:46
0:00:46