filmov
tv
How to Declare a List in Python
Показать описание
In this video, I will explain what is a list data structure and how a list is declared in Python language.
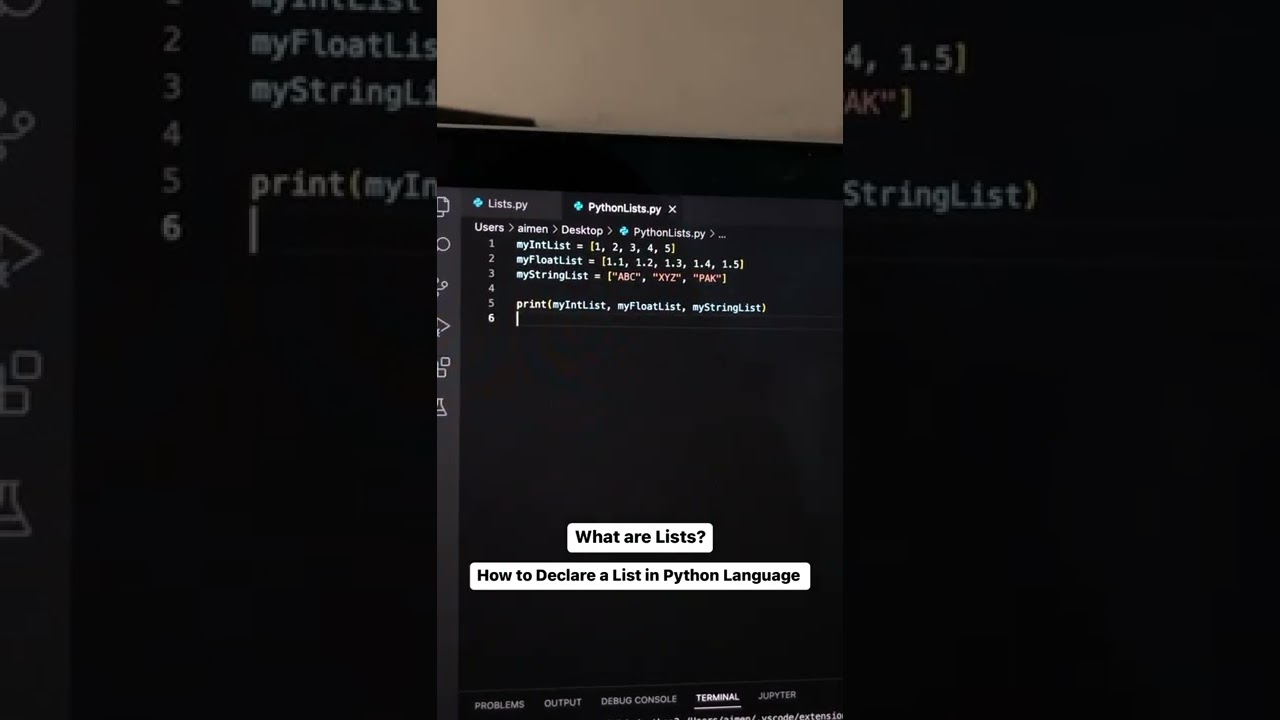
Lists are used to store multiple items in a single variable. Lists are used to store data in a sequential way. In Python, various built-in functions can be utilized for quick data manipulation and analysis. A list is declared within a bracket and it can store an integer, a float, or a string. The elements in a list are separated by a comma.
Here, we have declared three different types of lists. The first list contains integers, the second one contains floats, and the third one contains strings. After declaring these three lists we are printing them with the print command. After executing the print command, you can see the elements of each list being displayed in the terminal.
List items are ordered, changeable, and allow duplicate values. List items are indexed, the first item has an index [0], the second item has an index [1], and so on. When we say that lists are ordered, it means that the items have a defined order, and that order will not change. If you add new items to a list, the new items will be placed at the end of the list. The list is changeable, meaning that we can change, add, and remove items in a list after it has been created.
Join BioCode’s Advanced Bioinformatics Scripting in Python, BioPython, R & BioConductor course to intensify your biological programming career by learning through various useful & informative pre-recorded lectures on various biological programming/scripting languages.
This course includes:
-Introduction to Python, BioPython, R, Linux & BioConductor
-BLAST Database Searching, Parsing, and Extraction
-Sequence Analysis, Sequence Data Parsing, Sequence Retrieval, and Alignment
-Phylogenetic Analysis
-Processing and Analysis of Biological Datasets
-Data Visualization: ggplot2
-Bioinformatics File Parsing and Writing
-Gene Enrichment Analysis
-MicroArray Analysis: BioConductor
-RNA-Seq Analysis
-Variant Calling
To learn more about Python DM us, we can help you get started. BioCode provides an interactive platform to learn biological programming in Python & R, bioinformatics techniques, tools, databases, and biological data analysis in a cooperative manner covering both theoretical and practical aspects of computational biology topics. BioCode provides you with videos regarding every topic along with exercises. BioCode allows you to learn at your pace according to your schedule. Along with every video, BioCode provides you with transcriptions and PowerPoint presentations regarding that topic. If you have any queries during the lectures, there’s a dedicated section available for you to ask questions from your tutor.
#bioinformatics #computationalbiology #datascience #biology #biotechnology #scripting #coding #molecularbiology #programming #learncode #datavisualization #dataanalysis #drugdesigning #science #evolution #learn #biochemistry #microbiology #zoology #courses #python #immunology #shorts
Lists are used to store multiple items in a single variable. Lists are used to store data in a sequential way. In Python, various built-in functions can be utilized for quick data manipulation and analysis. A list is declared within a bracket and it can store an integer, a float, or a string. The elements in a list are separated by a comma.
Here, we have declared three different types of lists. The first list contains integers, the second one contains floats, and the third one contains strings. After declaring these three lists we are printing them with the print command. After executing the print command, you can see the elements of each list being displayed in the terminal.
List items are ordered, changeable, and allow duplicate values. List items are indexed, the first item has an index [0], the second item has an index [1], and so on. When we say that lists are ordered, it means that the items have a defined order, and that order will not change. If you add new items to a list, the new items will be placed at the end of the list. The list is changeable, meaning that we can change, add, and remove items in a list after it has been created.
Join BioCode’s Advanced Bioinformatics Scripting in Python, BioPython, R & BioConductor course to intensify your biological programming career by learning through various useful & informative pre-recorded lectures on various biological programming/scripting languages.
This course includes:
-Introduction to Python, BioPython, R, Linux & BioConductor
-BLAST Database Searching, Parsing, and Extraction
-Sequence Analysis, Sequence Data Parsing, Sequence Retrieval, and Alignment
-Phylogenetic Analysis
-Processing and Analysis of Biological Datasets
-Data Visualization: ggplot2
-Bioinformatics File Parsing and Writing
-Gene Enrichment Analysis
-MicroArray Analysis: BioConductor
-RNA-Seq Analysis
-Variant Calling
To learn more about Python DM us, we can help you get started. BioCode provides an interactive platform to learn biological programming in Python & R, bioinformatics techniques, tools, databases, and biological data analysis in a cooperative manner covering both theoretical and practical aspects of computational biology topics. BioCode provides you with videos regarding every topic along with exercises. BioCode allows you to learn at your pace according to your schedule. Along with every video, BioCode provides you with transcriptions and PowerPoint presentations regarding that topic. If you have any queries during the lectures, there’s a dedicated section available for you to ask questions from your tutor.
#bioinformatics #computationalbiology #datascience #biology #biotechnology #scripting #coding #molecularbiology #programming #learncode #datavisualization #dataanalysis #drugdesigning #science #evolution #learn #biochemistry #microbiology #zoology #courses #python #immunology #shorts
 0:17:36
0:17:36
 0:04:47
0:04:47
 0:13:08
0:13:08
 0:25:27
0:25:27
 0:11:57
0:11:57
 0:21:36
0:21:36
 0:05:15
0:05:15
 0:07:43
0:07:43
 0:00:32
0:00:32
 0:09:59
0:09:59
 0:06:26
0:06:26
 0:13:26
0:13:26
 0:08:18
0:08:18
 0:29:01
0:29:01
 0:13:58
0:13:58
 0:10:10
0:10:10
 0:23:47
0:23:47
 0:03:57
0:03:57
 0:06:15
0:06:15
 0:36:39
0:36:39
 0:00:35
0:00:35
 0:08:56
0:08:56
 0:09:14
0:09:14
 1:27:24
1:27:24